Characterization of the tandem CWCH2 sequence motif: a hallmark of inter-zinc finger interactions
- PMID: 20167128
- PMCID: PMC2837044
- DOI: 10.1186/1471-2148-10-53
Characterization of the tandem CWCH2 sequence motif: a hallmark of inter-zinc finger interactions
Abstract
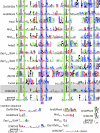
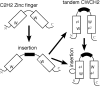
Background: The C2H2 zinc finger (ZF) domain is widely conserved among eukaryotic proteins. In Zic/Gli/Zap1 C2H2 ZF proteins, the two N-terminal ZFs form a single structural unit by sharing a hydrophobic core. This structural unit defines a new motif comprised of two tryptophan side chains at the center of the hydrophobic core. Because each tryptophan residue is located between the two cysteine residues of the C2H2 motif, we have named this structure the tandem CWCH2 (tCWCH2) motif.
Results: Here, we characterized 587 tCWCH2-containing genes using data derived from public databases. We categorized genes into 11 classes including Zic/Gli/Glis, Arid2/Rsc9, PacC, Mizf, Aebp2, Zap1/ZafA, Fungl, Zfp106, Twincl, Clr1, and Fungl-4ZF, based on sequence similarity, domain organization, and functional similarities. tCWCH2 motifs are mostly found in organisms belonging to the Opisthokonta (metazoa, fungi, and choanoflagellates) and Amoebozoa (amoeba, Dictyostelium discoideum). By comparison, the C2H2 ZF motif is distributed widely among the eukaryotes. The structure and organization of the tCWCH2 motif, its phylogenetic distribution, and molecular phylogenetic analysis suggest that prototypical tCWCH2 genes existed in the Opisthokonta ancestor. Within-group or between-group comparisons of the tCWCH2 amino acid sequence identified three additional sequence features (site-specific amino acid frequencies, longer linker sequence between two C2H2 ZFs, and frequent extra-sequences within C2H2 ZF motifs).
Conclusion: These features suggest that the tCWCH2 motif is a specialized motif involved in inter-zinc finger interactions.
Figures




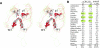

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

