Rational design of interleukin-21 antagonist through selective elimination of the gammaC binding epitope
- PMID: 20167599
- PMCID: PMC2852961
- DOI: 10.1074/jbc.M110.101444
Rational design of interleukin-21 antagonist through selective elimination of the gammaC binding epitope
Abstract
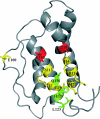
The cytokine interleukin (IL)-21 exerts pleiotropic effects acting through innate as well as adaptive immune responses. The activities of IL-21 are mediated through binding to its cognate receptor complex composed of the IL-21 receptor private chain (IL-21Ralpha) and the common gamma-chain (gammaC), the latter being shared by IL-2, IL-4, IL-7, IL-9, and IL-15. The binding energy of the IL-21 ternary complex is predominantly provided by the high affinity interaction between IL-21 and IL-21Ralpha, whereas the interaction between IL-21 and gammaC, albeit essential for signaling, is rather weak. The design of IL-21 analogues, which have lost most or all affinity toward the signaling gammaC chain, while simultaneously maintaining a tight interaction with the private chain, would in theory represent candidates for IL-21 antagonists. We predicted the IL-21 residues, which compose the gammaC binding epitope using homology modeling and alignment with the related cytokines, IL-2 and IL-4. Next we systematically analyzed the predicted binding epitope by a mutagenesis study. Indeed two mutants, which have significantly impaired gammaC affinity with undiminished IL-21Ralpha affinity, were successfully identified. Functional studies confirmed that these two novel hIL-21 double mutants do act as hIL-21 antagonists.
Figures

Similar articles
-
Crystal structure of interleukin-21 receptor (IL-21R) bound to IL-21 reveals that sugar chain interacting with WSXWS motif is integral part of IL-21R.J Biol Chem. 2012 Mar 16;287(12):9454-60. doi: 10.1074/jbc.M111.311084. Epub 2012 Jan 10. J Biol Chem. 2012. PMID: 22235133 Free PMC article.
-
Human IL-21 and IL-4 bind to partially overlapping epitopes of common gamma-chain.Biochem Biophys Res Commun. 2003 Jan 10;300(2):291-6. doi: 10.1016/s0006-291x(02)02836-x. Biochem Biophys Res Commun. 2003. PMID: 12504082
-
Three loops of the common gamma chain ectodomain required for the binding of interleukin-2 and interleukin-7.J Biol Chem. 2000 Sep 29;275(39):30100-5. doi: 10.1074/jbc.M004976200. J Biol Chem. 2000. PMID: 10887198
-
Structure, binding, and antagonists in the IL-4/IL-13 receptor system.Biochim Biophys Acta. 2002 Nov 11;1592(3):237-50. doi: 10.1016/s0167-4889(02)00318-x. Biochim Biophys Acta. 2002. PMID: 12421669 Review.
-
Structural Basis for Signaling Through Shared Common γ Chain Cytokines.Adv Exp Med Biol. 2019;1172:1-19. doi: 10.1007/978-981-13-9367-9_1. Adv Exp Med Biol. 2019. PMID: 31628649 Review.
Cited by
-
The Assessment of IL-21 and IL-22 at the mRNA Level in Tumor Tissue and Protein Concentration in Serum and Peritoneal Fluid in Patients with Ovarian Cancer.J Clin Med. 2021 Jul 9;10(14):3058. doi: 10.3390/jcm10143058. J Clin Med. 2021. PMID: 34300224 Free PMC article.
-
Generation and characterization of human anti-human IL-21 neutralizing monoclonal antibodies.MAbs. 2012 Jan-Feb;4(1):69-83. doi: 10.4161/mabs.4.1.18713. MAbs. 2012. PMID: 22327431 Free PMC article.
-
An HIV-1 envelope glycoprotein trimer with an embedded IL-21 domain activates human B cells.PLoS One. 2013 Jun 24;8(6):e67309. doi: 10.1371/journal.pone.0067309. Print 2013. PLoS One. 2013. PMID: 23826263 Free PMC article.
-
Crystal structure of interleukin-21 receptor (IL-21R) bound to IL-21 reveals that sugar chain interacting with WSXWS motif is integral part of IL-21R.J Biol Chem. 2012 Mar 16;287(12):9454-60. doi: 10.1074/jbc.M111.311084. Epub 2012 Jan 10. J Biol Chem. 2012. PMID: 22235133 Free PMC article.
-
Insights into cytokine-receptor interactions from cytokine engineering.Annu Rev Immunol. 2015;33:139-67. doi: 10.1146/annurev-immunol-032713-120211. Epub 2014 Dec 10. Annu Rev Immunol. 2015. PMID: 25493332 Free PMC article. Review.
References
-
- Spolski R., Leonard W. J. (2008) Annu. Rev. Immunol. 26, 57–79 - PubMed
-
- Nurieva R., Yang X. O., Martinez G., Zhang Y., Panopoulos A. D., Ma L., Schluns K., Tian Q., Watowich S. S., Jetten A. M., Dong C. (2007) Nature 448, 480–483 - PubMed
-
- Bondensgaard K., Breinholt J., Madsen D., Omkvist D. H., Kang L., Worsaae A., Becker P., Schiødt C. B., Hjorth S. A. (2007) J. Biol. Chem. 282, 23326–23336 - PubMed
-
- Asao H., Okuyama C., Kumaki S., Ishii N., Tsuchiya S., Foster D., Sugamura K. (2001) J. Immunol. 167, 1–5 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

