A new small regulatory protein, HmuP, modulates haemin acquisition in Sinorhizobium meliloti
- PMID: 20167620
- PMCID: PMC3068671
- DOI: 10.1099/mic.0.037713-0
A new small regulatory protein, HmuP, modulates haemin acquisition in Sinorhizobium meliloti
Abstract
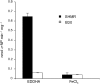
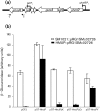
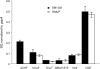
Sinorhizobium meliloti has multiple systems for iron acquisition, including the use of haem as an iron source. Haem internalization involves the ShmR haem outer membrane receptor and the hmuTUV locus, which participates in haem transport across the cytoplasmic membrane. Previous studies have demonstrated that expression of the shmR gene is negatively regulated by iron through RirA. Here, we identify hmuP in a genetic screen for mutants that displayed aberrant control of shmR. The normal induction of shmR in response to iron limitation was lost in the hmuP mutant, showing that this gene positively affects shmR expression. Moreover, the HmuP protein is not part of the haemin transporter system. Analysis of gene expression and siderophore production indicates that disruption of hmuP does not affect other genes related to the iron-restriction response. Our results strongly indicate that the main function of HmuP is the transcriptional regulation of shmR. Sequence alignment of HmuP homologues and comparison with the NMR structure of Rhodopseudomonas palustris CGA009 HmuP protein revealed that certain amino acids localized within predicted beta-sheets are well conserved. Our data indicate that at least one of the beta-sheets is important for HmuP activity.
Figures
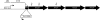

Similar articles
-
Highly conserved nucleotide motifs present in the 5'UTR of the heme-receptor gene shmR are required for HmuP-dependent expression of shmR in Ensifer meliloti.Biometals. 2019 Apr;32(2):273-291. doi: 10.1007/s10534-019-00184-6. Epub 2019 Feb 27. Biometals. 2019. PMID: 30810877
-
The hmuUV genes of Sinorhizobium meliloti 2011 encode the permease and ATPase components of an ABC transport system for the utilization of both haem and the hydroxamate siderophores, ferrichrome and ferrioxamine B.Mol Microbiol. 2008 Dec;70(5):1261-73. doi: 10.1111/j.1365-2958.2008.06479.x. Mol Microbiol. 2008. PMID: 18990190
-
ShmR is essential for utilization of heme as a nutritional iron source in Sinorhizobium meliloti.Appl Environ Microbiol. 2008 Oct;74(20):6473-5. doi: 10.1128/AEM.01590-08. Epub 2008 Aug 29. Appl Environ Microbiol. 2008. PMID: 18757569 Free PMC article.
-
Role of the regulatory gene rirA in the transcriptional response of Sinorhizobium meliloti to iron limitation.Appl Environ Microbiol. 2005 Oct;71(10):5969-82. doi: 10.1128/AEM.71.10.5969-5982.2005. Appl Environ Microbiol. 2005. PMID: 16204511 Free PMC article.
-
Identification of an iron-regulated, hemin-binding outer membrane protein in Sinorhizobium meliloti.Appl Environ Microbiol. 2002 Dec;68(12):5877-81. doi: 10.1128/AEM.68.12.5877-5881.2002. Appl Environ Microbiol. 2002. PMID: 12450806 Free PMC article.
Cited by
-
Differential control of Bradyrhizobium japonicum iron stimulon genes through variable affinity of the iron response regulator (Irr) for target gene promoters and selective loss of activator function.Mol Microbiol. 2014 May;92(3):609-24. doi: 10.1111/mmi.12584. Epub 2014 Apr 8. Mol Microbiol. 2014. PMID: 24646221 Free PMC article.
-
Methanotroph Methylotuvimicrobium alcaliphilum 20Z-3E as a fumarate producer: transcriptomic analysis and the role of malic enzyme.Int Microbiol. 2025 Mar 4. doi: 10.1007/s10123-025-00647-6. Online ahead of print. Int Microbiol. 2025. PMID: 40035991
-
Bradyrhizobium japonicum HmuP is an RNA-binding protein that positively controls hmuR operon expression by suppression of a negative regulatory RNA element in the 5' untranslated region.Mol Microbiol. 2024 Jun;121(6):1217-1227. doi: 10.1111/mmi.15274. Epub 2024 May 9. Mol Microbiol. 2024. PMID: 38725184 Free PMC article.
-
Transcriptomic response of Sinorhizobium meliloti to the predatory attack of Myxococcus xanthus.Front Microbiol. 2023 Jun 19;14:1213659. doi: 10.3389/fmicb.2023.1213659. eCollection 2023. Front Microbiol. 2023. PMID: 37405170 Free PMC article.
-
The Regulatory Protein ChuP Connects Heme and Siderophore-Mediated Iron Acquisition Systems Required for Chromobacterium violaceum Virulence.Front Cell Infect Microbiol. 2022 May 11;12:873536. doi: 10.3389/fcimb.2022.873536. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35646721 Free PMC article.
References
-
- Allaway, D., Schofield, N. A., Leonard, M. E., Gilardoni, L., Finan, T. M. & Poole, P. S. (2001). Use of differential fluorescence induction and optical trapping to isolate environmentally induced genes. Environ Microbiol 3, 397–406. - PubMed
-
- Andrews, S. C., Robinson, A. K. & Rodriguez-Quinones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol Rev 27, 215–237. - PubMed
-
- Arnold, K., Bordoli, L., Kopp, J. & Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. - PubMed
-
- Beringer, J. E. (1974). R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 84, 188–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

