Sent to destroy: the ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease
- PMID: 20167943
- PMCID: PMC2826711
- DOI: 10.1161/CIRCRESAHA.109.208801
Sent to destroy: the ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease
Abstract
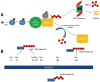
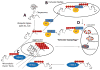
The ubiquitin proteasome system (UPS) plays a crucial role in biological processes integral to the development of the cardiovascular system and cardiovascular diseases. The UPS prototypically recognizes specific protein substrates and places polyubiquitin chains on them for subsequent destruction by the proteasome. This system is in place to degrade not only misfolded and damaged proteins, but is essential also in regulating a host of cell signaling pathways involved in proliferation, adaptation to stress, regulation of cell size, and cell death. During the development of the cardiovascular system, the UPS regulates cell signaling by modifying transcription factors, receptors, and structural proteins. Later, in the event of cardiovascular diseases as diverse as atherosclerosis, cardiac hypertrophy, and ischemia/reperfusion injury, ubiquitin ligases and the proteasome are implicated in protecting and exacerbating clinical outcomes. However, when misfolded and damaged proteins are ubiquitinated by the UPS, their destruction by the proteasome is not always possible because of their aggregated confirmations. Recent studies have discovered how these ubiquitinated misfolded proteins can be destroyed by alternative "specific" mechanisms. The cytosolic receptors p62, NBR, and histone deacetylase 6 recognize aggregated ubiquitinated proteins and target them for autophagy in the process of "selective autophagy." Even the ubiquitination of multiple proteins within whole organelles that drive the more general macro-autophagy may be due, in part, to similar ubiquitin-driven mechanisms. In summary, the crosstalk between the UPS and autophagy highlight the pivotal and diverse roles the UPS plays in maintaining protein quality control and regulating cardiovascular development and disease.
Figures

References
-
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8(6):610–616. - PubMed
-
- Leithe E, Rivedal E. Ubiquitination and down-regulation of gap junction protein connexin-43 in response to 12-O-tetradecanoylphorbol 13-acetate treatment. J Biol Chem. 2004;279(48):50089–50096. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL061656/HL/NHLBI NIH HHS/United States
- R01HL061656/HL/NHLBI NIH HHS/United States
- R37 HL065619/HL/NHLBI NIH HHS/United States
- R01 HL090823/HL/NHLBI NIH HHS/United States
- T32 HL069768/HL/NHLBI NIH HHS/United States
- P30 ES010126/ES/NIEHS NIH HHS/United States
- R01HL065619/HL/NHLBI NIH HHS/United States
- R01 HL065619/HL/NHLBI NIH HHS/United States
- R01 GM061728/GM/NIGMS NIH HHS/United States
- R01 HL104129/HL/NHLBI NIH HHS/United States
- R01GM061728/GM/NIGMS NIH HHS/United States
- T32 HL083828/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

