Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology
- PMID: 20168090
- PMCID: PMC2828578
- DOI: 10.4161/mabs.2.1.10788
Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology
Abstract
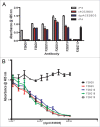
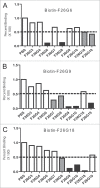
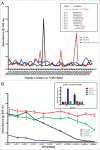
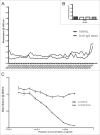
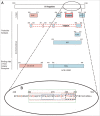
Neutralizing antibody responses to the surface glycoproteins of enveloped viruses play an important role in immunity. Many of these glycoproteins, including the severe acute respiratory syndrome-coronavirus (SARS-CoV) spike (S) protein form trimeric units in the membrane of the native virion. There is substantial experimental and pre-clinical evidence showing that the S protein is a promising lead for vaccines and therapeutics. Previously we generated a panel of monoclonal antibodies (mAbs) to whole inactivated SARS-CoV which neutralize the virus in vitro. Here, we define their specificity and affinity, map several of their epitopes and lastly characterise chimeric versions of them. Our data show that the neutralizing mAbs bind to the angiotensin-converting enzyme 2 (ACE2) receptor-binding domain (RBD) of the SARS S protein. Three of the chimeric mAbs retain their binding specificity while one conformational mAb, F26G19, lost its ability to bind the S protein despite high level expression. The affinity for recombinant S is maintained in all of the functional chimeric versions of the parental mAbs. Both parental mAb F26G18 and the chimeric version neutralize the TO R2 strain of SARS-CoV with essentially identical titres (2.07 and 2.47 nM, respectively). Lastly, a comparison with other neutralizing mAbs to SARS-CoV clearly shows that the dominance of a 33 amino acid residue loop of the SARS-CoV RBD is independent of repertoire, species, quaternary structure, and importantly, the technology used to derive the mAbs. In cases like this, the dominance of a compact RBD antigenic domain and the central role of the S protein in pathogenesis may inherently create immunoselection pressure on viruses to evolve more complex evasion strategies or die out of a host species. The apparent simplicity of the mechanism of SARS-CoV neutralization is in stark contrast to the complexity shown by other enveloped viruses.
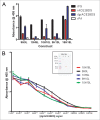
Figures
References
-
- Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
