Bacterial actin MreB assembles in complex with cell shape protein RodZ
- PMID: 20168300
- PMCID: PMC2845281
- DOI: 10.1038/emboj.2010.9
Bacterial actin MreB assembles in complex with cell shape protein RodZ
Abstract
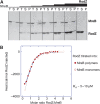
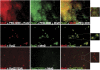
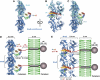
Bacterial actin homologue MreB is required for cell shape maintenance in most non-spherical bacteria, where it assembles into helical structures just underneath the cytoplasmic membrane. Proper assembly of the actin cytoskeleton requires RodZ, a conserved, bitopic membrane protein that colocalises to MreB and is essential for cell shape determination. Here, we present the first crystal structure of bacterial actin engaged with a natural partner and provide a clear functional significance of the interaction. We show that the cytoplasmic helix-turn-helix motif of Thermotoga maritima RodZ directly interacts with monomeric as well as filamentous MreB and present the crystal structure of the complex. In vitro and in vivo analyses of mutant T. maritima and Escherichia coli RodZ validate the structure and reveal the importance of the MreB-RodZ interaction in the ability of cells to propagate as rods. Furthermore, the results elucidate how the bacterial actin cytoskeleton might be anchored to the membrane to help constrain peptidoglycan synthesis in the periplasm.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli.EMBO J. 2009 Feb 4;28(3):193-204. doi: 10.1038/emboj.2008.264. Epub 2008 Dec 11. EMBO J. 2009. PMID: 19078962 Free PMC article.
-
Direct membrane binding by bacterial actin MreB.Mol Cell. 2011 Aug 5;43(3):478-87. doi: 10.1016/j.molcel.2011.07.008. Mol Cell. 2011. PMID: 21816350 Free PMC article.
-
The assembly of MreB, a prokaryotic homolog of actin.J Biol Chem. 2005 Jan 28;280(4):2628-35. doi: 10.1074/jbc.M410298200. Epub 2004 Nov 16. J Biol Chem. 2005. PMID: 15548516
-
The actin-like MreB proteins in Bacillus subtilis: a new turn.Front Biosci (Schol Ed). 2012 Jun 1;4(4):1582-606. doi: 10.2741/s354. Front Biosci (Schol Ed). 2012. PMID: 22652894 Review.
-
[Regulation of determination of bacterial shape].Nihon Saikingaku Zasshi. 2014;69(4):557-64. doi: 10.3412/jsb.69.557. Nihon Saikingaku Zasshi. 2014. PMID: 25447981 Review. Japanese.
Cited by
-
Tol-Pal System and Rgs Proteins Interact to Promote Unipolar Growth and Cell Division in Sinorhizobium meliloti.mBio. 2020 Jun 30;11(3):e00306-20. doi: 10.1128/mBio.00306-20. mBio. 2020. PMID: 32605980 Free PMC article.
-
YodL and YisK Possess Shape-Modifying Activities That Are Suppressed by Mutations in Bacillus subtilis mreB and mbl.J Bacteriol. 2016 Jul 13;198(15):2074-88. doi: 10.1128/JB.00183-16. Print 2016 Aug 1. J Bacteriol. 2016. PMID: 27215790 Free PMC article.
-
Roles of RodZ and class A PBP1b in the assembly and regulation of the peripheral peptidoglycan elongasome in ovoid-shaped cells of Streptococcus pneumoniae D39.Mol Microbiol. 2022 Oct;118(4):336-368. doi: 10.1111/mmi.14969. Epub 2022 Aug 24. Mol Microbiol. 2022. PMID: 36001060 Free PMC article.
-
Mechanical control of bacterial cell shape.Biophys J. 2011 Jul 20;101(2):327-35. doi: 10.1016/j.bpj.2011.06.005. Biophys J. 2011. PMID: 21767484 Free PMC article.
-
Proteomic analyses of nucleoid-associated proteins in Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus aureus.PLoS One. 2011 Apr 26;6(4):e19172. doi: 10.1371/journal.pone.0019172. PLoS One. 2011. PMID: 21541338 Free PMC article.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 - PubMed
-
- Aggarwal AK, Rodgers DW, Drottar M, Ptashne M, Harrison SC (1988) Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science 242: 899–907 - PubMed
-
- Amos LA, van den Ent F, Löwe J (2004) Structural/functional homology between the bacterial and eukaryotic cytoskeletons. Curr Opin Cell Biol 16: 24–31 - PubMed
-
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM (2005) The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29: 231–262 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

