Chromatin association and regulation of rDNA transcription by the Ras-family protein RasL11a
- PMID: 20168301
- PMCID: PMC2857460
- DOI: 10.1038/emboj.2010.16
Chromatin association and regulation of rDNA transcription by the Ras-family protein RasL11a
Abstract
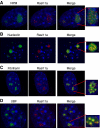
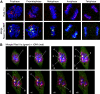
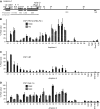
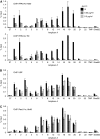
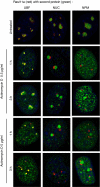
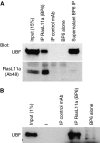
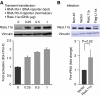
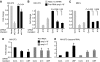
RasL11a and RasL11b are Ras super-family proteins of unknown function. Here, we show that RasL11a is a chromatin-associated modulator of pre-ribosomal RNA (pre-rRNA) synthesis. RasL11a was found in the nucleolus of interphase mouse fibroblasts, where it co-localized with the RNA polymerase I-specific transcription factor UBF. Similar to UBF, RasL11a also marked the active subset of rDNA repeats (also called nucleolar organizers, or NORs) on mitotic chromosomes. In cells, RasL11a existed in stable complexes with UBF and, as shown by chromatin immunoprecipitation, distributed along the rDNA transcription unit. Upon treatment of cells with actinomycin D, RasL11a and UBF persisted on the transcription unit beyond the release of RNA polymerase I, and remained co-localized in peri-nucleolar cap structures. Ectopic expression of RasL11a enhanced pre-rRNA levels in cells, whereas RasL11a knockdown had the opposite effect. In transient transfection experiments, RasL11a enhanced the transcriptional activity of an RNA polymerase I-specific reporter controlled by the rDNA enhancer/promoter region. We speculate that RasL11a acts in concert with UBF to facilitate initiation and/or elongation by RNA polymerase I in response to specific upstream stimuli.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
A unique enhancer boundary complex on the mouse ribosomal RNA genes persists after loss of Rrn3 or UBF and the inactivation of RNA polymerase I transcription.PLoS Genet. 2017 Jul 17;13(7):e1006899. doi: 10.1371/journal.pgen.1006899. eCollection 2017 Jul. PLoS Genet. 2017. PMID: 28715449 Free PMC article.
-
Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells.Genes Dev. 2007 Aug 15;21(16):2041-54. doi: 10.1101/gad.436707. Genes Dev. 2007. PMID: 17699751 Free PMC article.
-
Pseudo-NORs: a novel model for studying nucleoli.Biochim Biophys Acta. 2008 Nov;1783(11):2116-23. doi: 10.1016/j.bbamcr.2008.07.004. Epub 2008 Jul 18. Biochim Biophys Acta. 2008. PMID: 18687368 Review.
-
UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery.Genes Dev. 2005 Jan 1;19(1):50-64. doi: 10.1101/gad.310705. Epub 2004 Dec 14. Genes Dev. 2005. PMID: 15598984 Free PMC article.
-
A role for upstream binding factor in organizing ribosomal gene chromatin.Biochem Soc Symp. 2006;(73):77-84. doi: 10.1042/bss0730077. Biochem Soc Symp. 2006. PMID: 16626289 Review.
Cited by
-
H3K27me3 Does Not Orchestrate the Expression of Lineage-Specific Markers in hESC-Derived Hepatocytes In Vitro.Stem Cell Reports. 2016 Aug 9;7(2):192-206. doi: 10.1016/j.stemcr.2016.06.013. Epub 2016 Jul 28. Stem Cell Reports. 2016. PMID: 27477635 Free PMC article.
-
Dynamic regulation of EZH2 from HPSc to hepatocyte-like cell fate.PLoS One. 2017 Nov 1;12(11):e0186884. doi: 10.1371/journal.pone.0186884. eCollection 2017. PLoS One. 2017. PMID: 29091973 Free PMC article.
-
Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes.Subcell Biochem. 2013;61:211-36. doi: 10.1007/978-94-007-4525-4_10. Subcell Biochem. 2013. PMID: 23150253 Free PMC article. Review.
-
Spontaneous Tumor Regression in Tasmanian Devils Associated with RASL11A Activation.Genetics. 2020 Aug;215(4):1143-1152. doi: 10.1534/genetics.120.303428. Epub 2020 Jun 18. Genetics. 2020. PMID: 32554701 Free PMC article.
-
Induced Chromosomal Aneuploidy Results in Global and Consistent Deregulation of the Transcriptome of Cancer Cells.Neoplasia. 2019 Jul;21(7):721-729. doi: 10.1016/j.neo.2019.04.009. Epub 2019 Jun 4. Neoplasia. 2019. PMID: 31174021 Free PMC article.
References
-
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 - PubMed
-
- Bos JL, Rehmann H, Wittinghofer A (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129: 865–877 - PubMed
-
- Budde A, Grummt I (1999) p53 represses ribosomal gene transcription. Oncogene 18: 1119–1124 - PubMed
-
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW (2005) Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet 37: 1289–1295 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

