Structural basis for the function of DEAH helicases
- PMID: 20168331
- PMCID: PMC2838688
- DOI: 10.1038/embor.2010.11
Structural basis for the function of DEAH helicases
Abstract
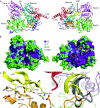
DEAH helicases participate in pre-messenger RNA splicing and ribosome biogenesis. The structure of yeast Prp43p-ADP reveals the homology of DEAH helicases to DNA helicases and the presence of an oligonucleotide-binding motif. A beta-hairpin from the second RecA domain is wedged between two carboxy-terminal domains and blocks access to the occluded RNA binding site formed by the RecA domains and a C-terminal domain. ATP binding and hydrolysis are likely to induce conformational changes in the hairpin that are important for RNA unwinding or ribonucleoprotein remodelling. The structure of Prp43p provides the framework for functional and genetic analysis of all DEAH helicases.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
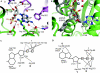

References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 - PubMed
-
- Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte MR (2004) Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol 11: 323–329 - PubMed
-
- Andersen CBF, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira C, Pedersen JS, Seraphin B, Le Hir H, Andersen GR (2006) Structure of the exon junction core complex with a trapped DEAD-Box ATPase bound to RNA. Science 313: 1968–1972 - PubMed
-
- Aravind L, Koonin EV (1999) G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem Sci 24: 342–344 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

