A class-wide phylogenetic assessment of Dothideomycetes
- PMID: 20169021
- PMCID: PMC2816964
- DOI: 10.3114/sim.2009.64.01
A class-wide phylogenetic assessment of Dothideomycetes
Abstract
We present a comprehensive phylogeny derived from 5 genes, nucSSU, nucLSU rDNA, TEF1, RPB1 and RPB2, for 356 isolates and 41 families (six newly described in this volume) in Dothideomycetes. All currently accepted orders in the class are represented for the first time in addition to numerous previously unplaced lineages. Subclass Pleosporomycetidae is expanded to include the aquatic order Jahnulales. An ancestral reconstruction of basic nutritional modes supports numerous transitions from saprobic life histories to plant associated and lichenised modes and a transition from terrestrial to aquatic habitats are confirmed. Finally, a genomic comparison of 6 dothideomycete genomes with other fungi finds a high level of unique protein associated with the class, supporting its delineation as a separate taxon.
Keywords: Ascomycota; Dothideomyceta; Pezizomycotina; Tree of Life; fungal evolution; lichens; multigene phylogeny; phylogenomics; plant pathogens; saprobes.
Figures

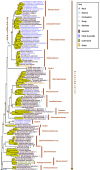



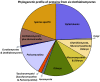
References
-
- Aptroot A, Lücking R, Sipman H, Umana L, Chaves J-L (2008). Pyrenocarpous lichens with bitunicate asci. A first assessment of the lichen biodiversity inventory in Costa Rica. Bibliotheca Lichenologica 97: 1–162.
-
- Arx von J, Müller E (1975). A re-evaluation of the bitunicate ascomycetes with keys to families and genera. Studies in Mycology 9: 1–159.
-
- Aveskamp MM, Verkley GJM, Gruyter J de, Murace MA, Perello A, et al. (2009). DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia 101: 359–378. - PubMed
-
- Barr ME (1979). A classification of Loculoascomycetes. Mycologia 71: 935–957.
-
- Barr ME (1987). Prodromus to class Loculoascomycetes. M.E. Barr Bigelow, Amherst, Massachusetts.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
