Visualizing interactions along the Escherichia coli twin-arginine translocation pathway using protein fragment complementation
- PMID: 20169075
- PMCID: PMC2821923
- DOI: 10.1371/journal.pone.0009225
Visualizing interactions along the Escherichia coli twin-arginine translocation pathway using protein fragment complementation
Abstract
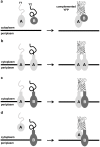
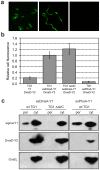
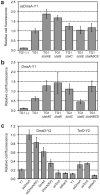
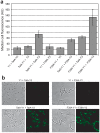
The twin-arginine translocation (Tat) pathway is well known for its ability to export fully folded substrate proteins out of the cytoplasm of gram-negative and gram-positive bacteria. Studies of this mechanism in Escherichia coli have identified numerous transient protein-protein interactions that guide export-competent proteins through the Tat pathway. To visualize these interactions, we have adapted bimolecular fluorescence complementation (BiFC) to detect protein-protein interactions along the Tat pathway of living cells. Fragments of the yellow fluorescent protein (YFP) were fused to soluble and transmembrane factors that participate in the translocation process including Tat substrates, Tat-specific proofreading chaperones and the integral membrane proteins TatABC that form the translocase. Fluorescence analysis of these YFP chimeras revealed a wide range of interactions such as the one between the Tat substrate dimethyl sulfoxide reductase (DmsA) and its dedicated proofreading chaperone DmsD. In addition, BiFC analysis illuminated homo- and hetero-oligomeric complexes of the TatA, TatB and TatC integral membrane proteins that were consistent with the current model of translocase assembly. In the case of TatBC assemblies, we provide the first evidence that these complexes are co-localized at the cell poles. Finally, we used this BiFC approach to capture interactions between the putative Tat receptor complex formed by TatBC and the DmsA substrate or its dedicated chaperone DmsD. Our results demonstrate that BiFC is a powerful approach for studying cytoplasmic and inner membrane interactions underlying bacterial secretory pathways.
Conflict of interest statement
Figures
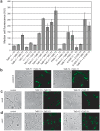
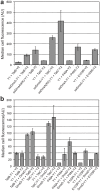
References
-
- Mori H, Ito K. The Sec protein-translocation pathway. Trends Microbiol. 2001;9:494–500. - PubMed
-
- Driessen AJ, Fekkes P, van der Wolk JP. The Sec system. Curr Opin Microbiol. 1998;1:216–22. - PubMed
-
- Driessen AJ, Manting EH, van der Does C. The structural basis of protein targeting and translocation in bacteria. Nat Struct Biol. 2001;8:492–8. - PubMed
-
- Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

