Inheritance of DNA transferred from American trypanosomes to human hosts
- PMID: 20169193
- PMCID: PMC2820539
- DOI: 10.1371/journal.pone.0009181
Inheritance of DNA transferred from American trypanosomes to human hosts
Abstract
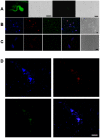
Interspecies DNA transfer is a major biological process leading to the accumulation of mutations inherited by sexual reproduction among eukaryotes. Lateral DNA transfer events and their inheritance has been challenging to document. In this study we modified a thermal asymmetric interlaced PCR by using additional targeted primers, along with Southern blots, fluorescence techniques, and bioinformatics, to identify lateral DNA transfer events from parasite to host. Instances of naturally occurring human infections by Trypanosoma cruzi are documented, where mitochondrial minicircles integrated mainly into retrotransposable LINE-1 of various chromosomes. The founders of five families show minicircle integrations that were transferred vertically to their progeny. Microhomology end-joining of 6 to 22 AC-rich nucleotide repeats in the minicircles and host DNA mediates foreign DNA integration. Heterogeneous minicircle sequences were distributed randomly among families, with diversity increasing due to subsequent rearrangement of inserted fragments. Mosaic recombination and hitchhiking on retrotransposition events to different loci were more prevalent in germ line as compared to somatic cells. Potential new genes, pseudogenes, and knockouts were identified. A pathway of minicircle integration and maintenance in the host genome is suggested. Thus, infection by T. cruzi has the unexpected consequence of increasing human genetic diversity, and Chagas disease may be a fortuitous share of negative selection. This demonstration of contemporary transfer of eukaryotic DNA to the human genome and its subsequent inheritance by descendants introduces a significant change in the scientific concept of evolutionary biology and medicine.
Conflict of interest statement
Figures
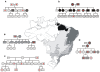
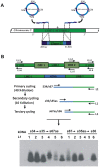
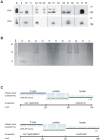
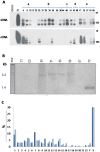

References
-
- WHO Control of Chagas disease: second report of a WHO Expert Committee. 2002. pp. 1–109.
-
- Teixeira AR, Nascimento R, Sturm NR. Evolution and Pathology in Chagas disease. Mem Inst Oswaldo Cruz. 2006;101:463–491. - PubMed
-
- Teixeira AR. Clinic Presentation of Chagas Disease. In: Teixeira A, Vinaud MC, Castro AM, editors. Emerging Chagas Disease. Bentham Science Publishers; 2009. pp. 104–109. : eISBN 978-1-60805-041-3.
-
- Teixeira AR, Figueiredo F, Rezende Filho J, Macedo V. Chagas disease: a clinical, parasitological and immunological study in rabbits. Am J Trop Med Hyg. 1983;32:258–272. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

