Mechanistic characterization and molecular modeling of hepatitis B virus polymerase resistance to entecavir
- PMID: 20169198
- PMCID: PMC2820545
- DOI: 10.1371/journal.pone.0009195
Mechanistic characterization and molecular modeling of hepatitis B virus polymerase resistance to entecavir
Abstract
Background: Entecavir (ETV) is a deoxyguanosine analog competitive inhibitor of hepatitis B virus (HBV) polymerase that exhibits delayed chain termination of HBV DNA. A high barrier to entecavir-resistance (ETVr) is observed clinically, likely due to its potency and a requirement for multiple resistance changes to overcome suppression. Changes in the HBV polymerase reverse-transcriptase (RT) domain involve lamivudine-resistance (LVDr) substitutions in the conserved YMDD motif (M204V/I +/- L180M), plus an additional ETV-specific change at residues T184, S202 or M250. These substitutions surround the putative dNTP binding site or primer grip regions of the HBV RT.
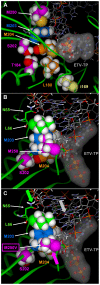
Methods/principal findings: To determine the mechanistic basis for ETVr, wildtype, lamivudine-resistant (M204V, L180M) and ETVr HBVs were studied using in vitro RT enzyme and cell culture assays, as well as molecular modeling. Resistance substitutions significantly reduced ETV incorporation and chain termination in HBV DNA and increased the ETV-TP inhibition constant (K(i)) for HBV RT. Resistant HBVs exhibited impaired replication in culture and reduced enzyme activity (k(cat)) in vitro. Molecular modeling of the HBV RT suggested that ETVr residue T184 was adjacent to and stabilized S202 within the LVDr YMDD loop. ETVr arose through steric changes at T184 or S202 or by disruption of hydrogen-bonding between the two, both of which repositioned the loop and reduced the ETV-triphosphate (ETV-TP) binding pocket. In contrast to T184 and S202 changes, ETVr at primer grip residue M250 was observed during RNA-directed DNA synthesis only. Experimentally, M250 changes also impacted the dNTP-binding site. Modeling suggested a novel mechanism for M250 resistance, whereby repositioning of the primer-template component of the dNTP-binding site shifted the ETV-TP binding pocket. No structural data are available to confirm the HBV RT modeling, however, results were consistent with phenotypic analysis of comprehensive substitutions of each ETVr position.
Conclusions: Altogether, ETVr occurred through exclusion of ETV-TP from the dNTP-binding site, through different, novel mechanisms that involved lamivudine-resistance, ETV-specific substitutions, and the primer-template.
Conflict of interest statement
Figures


Similar articles
-
Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.J Virol. 2021 Jul 26;95(16):e0240120. doi: 10.1128/JVI.02401-20. Epub 2021 Jul 26. J Virol. 2021. PMID: 34076480 Free PMC article.
-
Two-year assessment of entecavir resistance in Lamivudine-refractory hepatitis B virus patients reveals different clinical outcomes depending on the resistance substitutions present.Antimicrob Agents Chemother. 2007 Mar;51(3):902-11. doi: 10.1128/AAC.00833-06. Epub 2006 Dec 18. Antimicrob Agents Chemother. 2007. PMID: 17178796 Free PMC article. Clinical Trial.
-
Mechanism of entecavir resistance of hepatitis B virus with viral breakthrough as determined by long-term clinical assessment and molecular docking simulation.Antimicrob Agents Chemother. 2010 Feb;54(2):882-9. doi: 10.1128/AAC.01061-09. Epub 2009 Nov 23. Antimicrob Agents Chemother. 2010. PMID: 19933798 Free PMC article.
-
Structural basis of HIV inhibition by L-nucleosides: Opportunities for drug development and repurposing.Drug Discov Today. 2022 Jul;27(7):1832-1846. doi: 10.1016/j.drudis.2022.02.016. Epub 2022 Feb 23. Drug Discov Today. 2022. PMID: 35218925 Free PMC article. Review.
-
Entecavir: A Review and Considerations for Its Application in Oncology.Pharmaceuticals (Basel). 2023 Nov 14;16(11):1603. doi: 10.3390/ph16111603. Pharmaceuticals (Basel). 2023. PMID: 38004468 Free PMC article. Review.
Cited by
-
Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.J Virol. 2021 Jul 26;95(16):e0240120. doi: 10.1128/JVI.02401-20. Epub 2021 Jul 26. J Virol. 2021. PMID: 34076480 Free PMC article.
-
Modeling the functional state of the reverse transcriptase of hepatitis B virus and its application to probing drug-protein interaction.BMC Bioinformatics. 2016 Aug 31;17 Suppl 8(Suppl 8):280. doi: 10.1186/s12859-016-1116-4. BMC Bioinformatics. 2016. PMID: 27587008 Free PMC article.
-
Non-nucleoside hepatitis B virus polymerase inhibitors identified by an in vitro polymerase elongation assay.J Gastroenterol. 2020 Apr;55(4):441-452. doi: 10.1007/s00535-019-01643-0. Epub 2019 Nov 25. J Gastroenterol. 2020. PMID: 31768802
-
Differential binding of tenofovir and adefovir to reverse transcriptase of hepatitis B virus.PLoS One. 2014 Sep 2;9(9):e106324. doi: 10.1371/journal.pone.0106324. eCollection 2014. PLoS One. 2014. PMID: 25180507 Free PMC article.
-
Synthesis of Fluorinated Nucleosides/Nucleotides and Their Antiviral Properties.Molecules. 2024 May 19;29(10):2390. doi: 10.3390/molecules29102390. Molecules. 2024. PMID: 38792251 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

