The impact of genetic relationship information on genomic breeding values in German Holstein cattle
- PMID: 20170500
- PMCID: PMC2838754
- DOI: 10.1186/1297-9686-42-5
The impact of genetic relationship information on genomic breeding values in German Holstein cattle
Abstract
Background: The impact of additive-genetic relationships captured by single nucleotide polymorphisms (SNPs) on the accuracy of genomic breeding values (GEBVs) has been demonstrated, but recent studies on data obtained from Holstein populations have ignored this fact. However, this impact and the accuracy of GEBVs due to linkage disequilibrium (LD), which is fairly persistent over generations, must be known to implement future breeding programs.
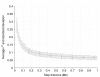
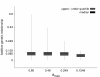
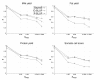
Materials and methods: The data set used to investigate these questions consisted of 3,863 German Holstein bulls genotyped for 54,001 SNPs, their pedigree and daughter yield deviations for milk yield, fat yield, protein yield and somatic cell score. A cross-validation methodology was applied, where the maximum additive-genetic relationship (amax) between bulls in training and validation was controlled. GEBVs were estimated by a Bayesian model averaging approach (BayesB) and an animal model using the genomic relationship matrix (G-BLUP). The accuracy of GEBVs due to LD was estimated by a regression approach using accuracy of GEBVs and accuracy of pedigree-based BLUP-EBVs.
Results: Accuracy of GEBVs obtained by both BayesB and G-BLUP decreased with decreasing amax for all traits analyzed. The decay of accuracy tended to be larger for G-BLUP and with smaller training size. Differences between BayesB and G-BLUP became evident for the accuracy due to LD, where BayesB clearly outperformed G-BLUP with increasing training size.
Conclusions: GEBV accuracy of current selection candidates varies due to different additive-genetic relationships relative to the training data. Accuracy of future candidates can be lower than reported in previous studies because information from close relatives will not be available when selection on GEBVs is applied. A Bayesian model averaging approach exploits LD information considerably better than G-BLUP and thus is the most promising method. Cross-validations should account for family structure in the data to allow for long-lasting genomic based breeding plans in animal and plant breeding.
Figures

References
-
- Malécot G. Les Mathématiques de l'Hérédité. Paris: Masson et Cie. vi + 1948. p. 63.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

