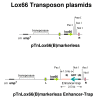
Replacing the wild type loxP site in BACs from the public domain with lox66 using a lox66 transposon
- PMID: 20170521
- PMCID: PMC2841073
- DOI: 10.1186/1756-0500-3-38
Replacing the wild type loxP site in BACs from the public domain with lox66 using a lox66 transposon
Abstract
Background: Chromatin adjoining the site of integration of a transgene affects expression and renders comparisons of closely related transgenes, such as those derived from a BAC deletion series retrofitted with enhancer-traps, unreliable. Gene targeting to a pre-determined site on the chromosome is likely to alleviate the problem.
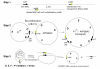
Findings: A general procedure to replace the loxP site located at one end of genomic DNA inserts in BACs with lox66 is described. Truncating insert DNA from the loxP end with a Tn10 transposon carrying a lox66 site simultaneously substitutes the loxP with a lox66 sequence. The replacement occurs with high stringency, and the procedure should be applicable to all BACs in the public domain. Cre recombination of loxP with lox66 or lox71 was found to be as efficient as another loxP site during phage P1 transduction of small plasmids containing those sites. However the end-deletion of insert DNA in BACs using a lox66 transposon occurred at no more than 20% the efficiency observed with a loxP transposon. Differences in the ability of Cre protein available at different stages of the P1 life cycle to recombine identical versus non-identical lox-sites is likely responsible for this discrepancy. A possible mechanism to explain these findings is discussed.
Conclusions: The loxP/lox66 replacement procedure should allow targeting BACs to a pre-positioned lox71 site in zebrafish chromosomes; a system where homologous recombination-mediated "knock-in" technology is unavailable.
Figures



Similar articles
-
Generating libraries of iTol2-end insertions at BAC ends using loxP and lox511 Tn10 transposons.BMC Genomics. 2011 Jul 7;12:351. doi: 10.1186/1471-2164-12-351. BMC Genomics. 2011. PMID: 21736732 Free PMC article.
-
Directing enhancer-traps and iTol2 end-sequences to deleted BAC ends with loxP- and lox511-Tn10 transposons.Methods Mol Biol. 2015;1227:99-122. doi: 10.1007/978-1-4939-1652-8_5. Methods Mol Biol. 2015. PMID: 25239743
-
A high-throughput screen identifying sequence and promiscuity characteristics of the loxP spacer region in Cre-mediated recombination.BMC Genomics. 2006 Apr 4;7:73. doi: 10.1186/1471-2164-7-73. BMC Genomics. 2006. PMID: 16595017 Free PMC article.
-
Isolating large nested deletions in bacterial and P1 artificial chromosomes by in vivo P1 packaging of products of Cre-catalysed recombination between the endogenous and a transposed loxP site.Nucleic Acids Res. 1997 Jun 1;25(11):2205-12. doi: 10.1093/nar/25.11.2205. Nucleic Acids Res. 1997. PMID: 9153322 Free PMC article.
-
Harnessing mobile genetic elements to explore gene regulation.Mob Genet Elements. 2014 Jul 7;4:e29759. doi: 10.4161/mge.29759. eCollection 2014. Mob Genet Elements. 2014. PMID: 25054085 Free PMC article. Review.
Cited by
-
Identifying Distal cis-acting Gene-Regulatory Sequences by Expressing BACs Functionalized with loxP-Tn10 Transposons in Zebrafish.RSC Adv. 2013 Jun 21;3(23):8604-8617. doi: 10.1039/C3RA40332G. RSC Adv. 2013. PMID: 24772295 Free PMC article.
-
Generating libraries of iTol2-end insertions at BAC ends using loxP and lox511 Tn10 transposons.BMC Genomics. 2011 Jul 7;12:351. doi: 10.1186/1471-2164-12-351. BMC Genomics. 2011. PMID: 21736732 Free PMC article.
-
Lox'd in translation: contradictions in the nomenclature surrounding common lox-site mutants and their implications in experiments.Microbiology (Reading). 2021 Jan;167(1):000997. doi: 10.1099/mic.0.000997. Epub 2020 Dec 7. Microbiology (Reading). 2021. PMID: 33284099 Free PMC article.
-
Site-specific integration of transgene targeting an endogenous lox-like site in early mouse embryos.J Appl Genet. 2011 Feb;52(1):89-94. doi: 10.1007/s13353-010-0011-3. Epub 2010 Nov 26. J Appl Genet. 2011. PMID: 21110150
References
-
- Sauer B, Hendenson N. Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. New Biol. 1990. pp. 441–449. - PubMed
LinkOut - more resources
Full Text Sources

