Multiplex nested RT-PCR for the detection of porcine enteric viruses
- PMID: 20170679
- PMCID: PMC7112813
- DOI: 10.1016/j.jviromet.2010.02.010
Multiplex nested RT-PCR for the detection of porcine enteric viruses
Abstract
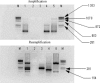
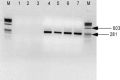
Porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), and porcine group A rotavirus (PRV-A) are major viruses causing enteric diseases of piglets. A multiplex nested reverse transcription polymerase chain reaction (multiplex nested RT-PCR) was developed for the detection of these viruses in field samples from piglets with diarrhea. A mixture of (1) three external pairs of primers, yielding in the amplification step two different amplicons with sizes of 950 bp and 317 bp and (2) three pairs of internal primers in a second round of PCR (nested PCR), yielding two different amplicons with sizes of 792 bp and 208 bp for TGEV and porcine PRV-A, respectively. The genome of PEDV was not detected after the amplification step but it was detected in the second round of PCR, yielding amplicon with size of 291 bp. Multiplex nested RT-PCR can detect TGEV, PRV-A, and PEDV up to concentration 10(2) TCID(50)/mL, 10(1) TCID(50)/mL, and 27.2 microg/microl of RNA, respectively. A total of 175 field samples were collected from swine with diarrhea from January 2005 until July 2007. The samples were tested for the presence of three viruses by a multiplex nested RT-PCR. Dual infections with PEDV and PRV-A were identified in seven specimens (4%) (n = 6). Twenty-one (25%) infections were caused by PEDV and thirty-four infections (41%) were caused by PRV-A. The genome of TGEV was not detected in any of these field samples, however TGEV was detected in piglets infected experimentally. The multiplex nested RT-PCR is rapid, sensitive, and a cost-effective detection method for the detection of porcine enteric viruses.
Copyright 2010 Elsevier B.V. All rights reserved.
Figures





References
-
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. - PubMed
-
- Dean A.G., Arner T.G., Sunki G.G., Friedman R., Lantinga M., Sangam S., Zubieta J.C., Sullivan K.M., Brendel K.A., Gao Z., Fontaine N., Shu M., Fuller G., Smith D.C., Nitschke D.A., Fagan R.F. Centers for Disease Control and Prevention; Atlanta, GA, USA: 2007. Epi Info™, A Database and Statistics Program for Public Health Professionals.
-
- Elschner M., Prudlo J., Hotzel H., Otto P., Sachse K. Nested reverse transcriptase polymerase chain reaction for the detection of group A rotavirus. J. Vet. Med. 2002;49:77–81. - PubMed
-
- Gribanov O.G., Shcherbakov A.V., Perevozchikova N.A., Drygin V.V., Gusev A.A. A simple method for RNA isolation and purification. Bioorg. Khim. 1997;23:763–765. (Abstract) - PubMed
-
- Hall T.A. BioEdit: a user friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999;41:95–98.
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

