Seoul virus suppresses NF-kappaB-mediated inflammatory responses of antigen presenting cells from Norway rats
- PMID: 20170933
- PMCID: PMC2837270
- DOI: 10.1016/j.virol.2010.01.027
Seoul virus suppresses NF-kappaB-mediated inflammatory responses of antigen presenting cells from Norway rats
Abstract
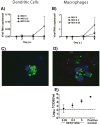
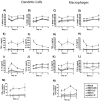
Hantavirus infection reduces antiviral defenses, increases regulatory responses, and causes persistent infection in rodent hosts. To address whether hantaviruses alter the maturation and functional activity of antigen presenting cells (APCs), rat bone marrow-derived dendritic cells (BMDCs) and macrophages (BMDMs) were generated and infected with Seoul virus (SEOV) or stimulated with TLR ligands. SEOV infected both DCs and macrophages, but copies of viral RNA, viral antigen, and infectious virus titers were higher in macrophages. The expression of MHCII and CD80, production of IL-6, IL-10, and TNF-alpha, and expression of Ifnbeta were attenuated in SEOV-infected APCs. Stimulation of APCs with poly I:C prior to SEOV infection increased the expression of activation markers and production of inflammatory cytokines and suppressed SEOV replication. Infection of APCs with SEOV suppressed LPS-induced activation and innate immune responses. Hantaviruses reduce the innate immune response potential of APCs derived from a natural host, which may influence persistence of these zoonotic viruses in the environment.
Copyright 2010 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures





Similar articles
-
Seoul virus enhances regulatory and reduces proinflammatory responses in male Norway rats.J Med Virol. 2008 Jul;80(7):1308-18. doi: 10.1002/jmv.21213. J Med Virol. 2008. PMID: 18461618 Free PMC article.
-
Seoul virus-infected rat lung endothelial cells and alveolar macrophages differ in their ability to support virus replication and induce regulatory T cell phenotypes.J Virol. 2012 Nov;86(21):11845-55. doi: 10.1128/JVI.01233-12. Epub 2012 Aug 22. J Virol. 2012. PMID: 22915818 Free PMC article.
-
Seoul orthohantavirus evades innate immune activation by reservoir endothelial cells.PLoS Pathog. 2024 Nov 25;20(11):e1012728. doi: 10.1371/journal.ppat.1012728. eCollection 2024 Nov. PLoS Pathog. 2024. PMID: 39585900 Free PMC article.
-
Infection, modulation and responses of antigen-presenting cells to African swine fever viruses.Virus Res. 2018 Oct 15;258:73-80. doi: 10.1016/j.virusres.2018.10.007. Epub 2018 Oct 11. Virus Res. 2018. PMID: 30316802 Review.
-
Role of Seaports and Imported Rats in Seoul Hantavirus Circulation, Africa.Emerg Infect Dis. 2023 Jan;29(1):20-25. doi: 10.3201/eid2901.221092. Emerg Infect Dis. 2023. PMID: 36573519 Free PMC article. Review.
Cited by
-
Species-specific responses during Seoul orthohantavirus infection in human and rat lung microvascular endothelial cells.PLoS Negl Trop Dis. 2024 Mar 27;18(3):e0012074. doi: 10.1371/journal.pntd.0012074. eCollection 2024 Mar. PLoS Negl Trop Dis. 2024. PMID: 38536871 Free PMC article.
-
Increased blood CD226- inflammatory monocytes with low antigen presenting potential correlate positively with severity of hemorrhagic fever with renal syndrome.Ann Med. 2023;55(2):2247000. doi: 10.1080/07853890.2023.2247000. Ann Med. 2023. PMID: 37585670 Free PMC article.
-
Puumala orthohantavirus: prevalence, biology, disease, animal models and recent advances in therapeutics development and structural biology.Front Immunol. 2025 May 8;16:1575112. doi: 10.3389/fimmu.2025.1575112. eCollection 2025. Front Immunol. 2025. PMID: 40406115 Free PMC article. Review.
-
Hantavirus immunology of rodent reservoirs: current status and future directions.Viruses. 2014 Mar 14;6(3):1317-35. doi: 10.3390/v6031317. Viruses. 2014. PMID: 24638205 Free PMC article. Review.
-
Innate Immunity to Orthohantaviruses: Could Divergent Immune Interactions Explain Host-specific Disease Outcomes?J Mol Biol. 2022 Mar 30;434(6):167230. doi: 10.1016/j.jmb.2021.167230. Epub 2021 Sep 4. J Mol Biol. 2022. PMID: 34487792 Free PMC article. Review.
References
-
- Beck K, Meyer-Konig U, Weidmann M, Nern C, Hufert FT. Human cytomegalovirus impairs dendritic cell function: a novel mechanism of human cytomegalovirus immune escape. Eur J Immunol. 2003;33:1528–1538. - PubMed
-
- Boltz-Nitulescu G, Wiltschke C, Holzinger C, Fellinger A, Scheiner O, Gessl A, Forster O. Differentiation of rat bone marrow cells into macrophages under the influence of mouse L929 cell supernatant. J Leukoc Biol. 1987;41:83–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

