An efficient approach to isolate STAT regulated enhancers uncovers STAT92E fundamental role in Drosophila tracheal development
- PMID: 20171201
- PMCID: PMC2877871
- DOI: 10.1016/j.ydbio.2010.02.015
An efficient approach to isolate STAT regulated enhancers uncovers STAT92E fundamental role in Drosophila tracheal development
Abstract
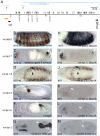
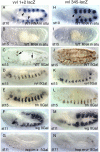
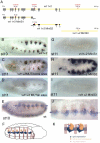
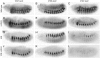
The ventral veinless (vvl) and trachealess (trh) genes are determinants of the Drosophila trachea. Early in development both genes are independently activated in the tracheal primordia by signals that are ill defined. Mutants blocking JAK/STAT signaling at any level do not form a tracheal tree suggesting that STAT92E may be an upstream transcriptional activator of the early trachea determinants. To test this hypothesis we have searched for STAT92E responsive enhancers activating the expression of vvl and trh in the tracheal primordia. We show that STAT92E regulated enhancers can be rapidly and efficiently isolated by focusing the analysis on genomic regions with clusters of putative STAT binding sites where at least some of them are phylogenetically conserved. Detailed analysis of a vvl early tracheal enhancer shows that non-conserved sites collaborate with conserved sites for enhancer activation. We find that STAT92E regulated enhancers can be located as far 60 kb from the promoters. Our results indicate that vvl and trh are independently activated by STAT92E which is the most important transcription factor required for trachea specification.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Trachealess (Trh) regulates all tracheal genes during Drosophila embryogenesis.Dev Biol. 2011 Dec 1;360(1):160-72. doi: 10.1016/j.ydbio.2011.09.014. Epub 2011 Sep 22. Dev Biol. 2011. PMID: 21963537 Free PMC article.
-
ventral veinless, a POU domain transcription factor, regulates different transduction pathways required for tracheal branching in Drosophila.Development. 1997 Sep;124(17):3273-81. doi: 10.1242/dev.124.17.3273. Development. 1997. PMID: 9310322
-
GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo.Gene Expr Patterns. 2007 Jan;7(3):323-31. doi: 10.1016/j.modgep.2006.08.003. Epub 2006 Aug 22. Gene Expr Patterns. 2007. PMID: 17008134
-
Chemical Embryology Redux: Metabolic Control of Development.Trends Genet. 2020 Aug;36(8):577-586. doi: 10.1016/j.tig.2020.05.007. Epub 2020 Jun 9. Trends Genet. 2020. PMID: 32532533 Free PMC article. Review.
-
The tracheal immune system of insects - A blueprint for understanding epithelial immunity.Insect Biochem Mol Biol. 2023 Jun;157:103960. doi: 10.1016/j.ibmb.2023.103960. Epub 2023 May 24. Insect Biochem Mol Biol. 2023. PMID: 37235953 Review.
Cited by
-
Development and Function of the Drosophila Tracheal System.Genetics. 2018 Jun;209(2):367-380. doi: 10.1534/genetics.117.300167. Genetics. 2018. PMID: 29844090 Free PMC article. Review.
-
In vivo Hox binding specificity revealed by systematic changes to a single cis regulatory module.Nat Commun. 2019 Aug 9;10(1):3597. doi: 10.1038/s41467-019-11416-1. Nat Commun. 2019. PMID: 31399572 Free PMC article.
-
Trachealess (Trh) regulates all tracheal genes during Drosophila embryogenesis.Dev Biol. 2011 Dec 1;360(1):160-72. doi: 10.1016/j.ydbio.2011.09.014. Epub 2011 Sep 22. Dev Biol. 2011. PMID: 21963537 Free PMC article.
-
Genome-wide identification of Grainy head targets in Drosophila reveals regulatory interactions with the POU domain transcription factor Vvl.Development. 2017 Sep 1;144(17):3145-3155. doi: 10.1242/dev.143297. Epub 2017 Jul 31. Development. 2017. PMID: 28760809 Free PMC article.
-
Ribbon boosts ribosomal protein gene expression to coordinate organ form and function.J Cell Biol. 2022 Apr 4;221(4):e202110073. doi: 10.1083/jcb.202110073. Epub 2022 Feb 23. J Cell Biol. 2022. PMID: 35195669 Free PMC article.
References
-
- Anderson M.G., Certel S.J., Certel K., Lee T., Montell D.J., Johnson W.A. Function of the Drosophila POU domain transcription factor drifter as an upstream regulator of breathless receptor tyrosine kinase expression in developing trachea. Development. 1996;122:4169–4178. - PubMed
-
- Anderson M.G., Perkins G.L., Chittick P., Shrigley R.J., Johnson W.A. drifter, a Drosophila POU-domain transcription factor, is required for correct differentiation and migration of tracheal cells and midline glia. Genes Dev. 1995;9:123–137. - PubMed
-
- Arbouzova N.I., Zeidler M.P. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–2616. - PubMed
-
- Bergad P.L., Shih H.M., Towle H.C., Schwarzenberg S.J., Berry S.A. Growth hormone induction of hepatic serine protease inhibitor 2.1 transcription is mediated by a Stat5-related factor binding synergistically to two gamma-activated sites. J. Biol. Chem. 1995;270:24903–24910. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

