In silico characterization and homology modeling of thylakoid-bound ascorbate peroxidase from a drought tolerant wheat cultivar
- PMID: 20172491
- PMCID: PMC5054410
- DOI: 10.1016/S1672-0229(08)60048-0
In silico characterization and homology modeling of thylakoid-bound ascorbate peroxidase from a drought tolerant wheat cultivar
Abstract
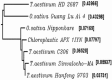
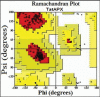
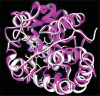
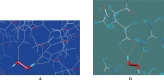
Ascorbate peroxidase, a haem protein (EC 1.11.1.11), efficiently scavenges hydrogen peroxide (H(2)O(2)) in cytosol and chloroplasts of plants. In this study, a full-length coding sequence of thylakoid-bound ascorbate peroxidase cDNA (TatAPX) was cloned from a drought tolerant wheat cultivar C306. Homology modeling of the TatAPX protein was performed by using the template crystal structure of chloroplastic ascorbate peroxidase from tobacco plant (PDB: 1IYN). The model structure was further refined by molecular mechanics and dynamic methods using various tools such as PROCHECK, ProSA and Verify3D. The predicted model was then tested for docking with H(2)O(2), the substrate for TatAPX enzyme. The results revealed that Arg233 and Glu255 in the predicted active site of the enzyme are two important amino acid residues responsible for strong hydrogen bonding affinity with H(2)O(2), which might play an important role in scavenging of H(2)O(2) from the plant system.
Copyright 2009 Beijing Genomics Institute. Published by Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
cDNA cloning of thylakoid-bound ascorbate peroxidase in pumpkin and its characterization.Plant Cell Physiol. 1996 Apr;37(3):405-9. doi: 10.1093/oxfordjournals.pcp.a028961. Plant Cell Physiol. 1996. PMID: 8673346
-
An inserted loop region of stromal ascorbate peroxidase is involved in its hydrogen peroxide-mediated inactivation.FEBS J. 2006 Jun;273(12):2704-10. doi: 10.1111/j.1742-4658.2006.05286.x. FEBS J. 2006. PMID: 16817898
-
From sequence analysis of three novel ascorbate peroxidases from Arabidopsis thaliana to structure, function and evolution of seven types of ascorbate peroxidase.Biochem J. 1997 Sep 1;326 ( Pt 2)(Pt 2):305-10. doi: 10.1042/bj3260305. Biochem J. 1997. PMID: 9291097 Free PMC article.
-
Hydrogen peroxide-mediated inactivation of two chloroplastic peroxidases, ascorbate peroxidase and 2-cys peroxiredoxin.Photochem Photobiol. 2008 Nov-Dec;84(6):1404-9. doi: 10.1111/j.1751-1097.2008.00452.x. Photochem Photobiol. 2008. PMID: 19067962 Review.
-
Chloroplastic ascorbate peroxidases targeted to stroma or thylakoid membrane: The chicken or egg dilemma.FEBS Lett. 2022 Dec;596(23):2989-3004. doi: 10.1002/1873-3468.14438. Epub 2022 Aug 19. FEBS Lett. 2022. PMID: 35776057 Review.
Cited by
-
Phylogenetic analyses, protein modeling and active site prediction of two pathogenesis related (PR2 and PR3) genes from bread wheat.PLoS One. 2021 Sep 10;16(9):e0257392. doi: 10.1371/journal.pone.0257392. eCollection 2021. PLoS One. 2021. PMID: 34506613 Free PMC article.
-
Real-Time Determination of Intracellular cAMP Reveals Functional Coupling of Gs Protein to the Melatonin MT1 Receptor.Int J Mol Sci. 2024 Mar 2;25(5):2919. doi: 10.3390/ijms25052919. Int J Mol Sci. 2024. PMID: 38474167 Free PMC article.
-
Molecular cloning and in-silico characterization of high temperature stress responsive pAPX gene isolated from heat tolerant Indian wheat cv. Raj 3765.BMC Res Notes. 2014 Oct 10;7:713. doi: 10.1186/1756-0500-7-713. BMC Res Notes. 2014. PMID: 25304397 Free PMC article.
References
-
- Hong C.Y. Expression of ascorbate peroxidase 8 in roots of rice (Oryza sativa L.) seedlings in response to NaCl. J. Exp. Bot. 2007;58:3273–3283. - PubMed
-
- Apel K., Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant. Biol. 2004;55:373–399. - PubMed
-
- Asada K. Ascorbate peroxidase—a hydrogen peroxide-scavenging enzyme in plants. Physiol. Plant. 1992;85:235–241.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

