The capsid-spacer peptide 1 Gag processing intermediate is a dominant-negative inhibitor of HIV-1 maturation
- PMID: 20172577
- PMCID: PMC2838455
- DOI: 10.1016/j.virol.2010.01.028
The capsid-spacer peptide 1 Gag processing intermediate is a dominant-negative inhibitor of HIV-1 maturation
Abstract
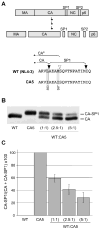
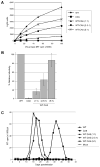
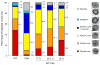
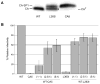
The human immunodeficiency virus type 1 (HIV-1) maturation inhibitor bevirimat disrupts virus replication by inhibiting the cleavage of the capsid-spacer peptide 1 (CA-SP1) Gag processing intermediate to mature CA. The observation that bevirimat delays but does not completely block CA-SP1 processing suggests that the presence of uncleaved CA-SP1 may disrupt the maturation process in trans. In this study, we validate this hypothesis by using a genetic approach to demonstrate that a non-cleavable CA-SP1 mutant exerts a dominant-negative effect on maturation of wild-type HIV-1. In contrast, a mutant in which cleavage can occur internally within SP1 is significantly less potent as a dominant-negative inhibitor. We also show that bevirimat blocks processing at both the major CA-SP1 cleavage site and the internal site. These data underscore the importance of full CA-SP1 processing for HIV-1 maturation and highlight the therapeutic potential of inhibitors that target this Gag cleavage event.
Published by Elsevier Inc.
Figures
Similar articles
-
Resistance to Second-Generation HIV-1 Maturation Inhibitors.J Virol. 2019 Mar 5;93(6):e02017-18. doi: 10.1128/JVI.02017-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30567982 Free PMC article.
-
Alkyl Amine Bevirimat Derivatives Are Potent and Broadly Active HIV-1 Maturation Inhibitors.Antimicrob Agents Chemother. 2015 Oct 19;60(1):190-7. doi: 10.1128/AAC.02121-15. Print 2016 Jan. Antimicrob Agents Chemother. 2015. PMID: 26482309 Free PMC article.
-
The prototype HIV-1 maturation inhibitor, bevirimat, binds to the CA-SP1 cleavage site in immature Gag particles.Retrovirology. 2011 Dec 7;8:101. doi: 10.1186/1742-4690-8-101. Retrovirology. 2011. PMID: 22151792 Free PMC article.
-
HIV-1 Maturation: Lessons Learned from Inhibitors.Viruses. 2020 Aug 26;12(9):940. doi: 10.3390/v12090940. Viruses. 2020. PMID: 32858867 Free PMC article. Review.
-
Bevirimat: a novel maturation inhibitor for the treatment of HIV-1 infection.Antivir Chem Chemother. 2008;19(3):107-13. doi: 10.1177/095632020801900301. Antivir Chem Chemother. 2008. PMID: 19024627 Review.
Cited by
-
My Cousin, My Enemy: quasispecies suppression of drug resistance.Curr Opin Virol. 2016 Oct;20:106-111. doi: 10.1016/j.coviro.2016.09.011. Epub 2016 Oct 17. Curr Opin Virol. 2016. PMID: 27764731 Free PMC article. Review.
-
Preservation of HIV-1 Gag Helical Bundle Symmetry by Bevirimat Is Central to Maturation Inhibition.J Am Chem Soc. 2021 Nov 17;143(45):19137-19148. doi: 10.1021/jacs.1c08922. Epub 2021 Nov 5. J Am Chem Soc. 2021. PMID: 34739240 Free PMC article.
-
Identification of an HIV-1 Mutation in Spacer Peptide 1 That Stabilizes the Immature CA-SP1 Lattice.J Virol. 2015 Nov 4;90(2):972-8. doi: 10.1128/JVI.02204-15. Print 2016 Jan 15. J Virol. 2015. PMID: 26537676 Free PMC article.
-
Maturation of the HIV-1 core by a non-diffusional phase transition.Nat Commun. 2015 Jan 8;6:5854. doi: 10.1038/ncomms6854. Nat Commun. 2015. PMID: 25569620 Free PMC article.
-
A Truncated Nef Peptide from SIVcpz Inhibits the Production of HIV-1 Infectious Progeny.Viruses. 2016 Jul 7;8(7):189. doi: 10.3390/v8070189. Viruses. 2016. PMID: 27399760 Free PMC article.
References
-
- Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv Pharmacol. 2007;55:347–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

