PB1-F2 influenza A virus protein adopts a beta-sheet conformation and forms amyloid fibers in membrane environments
- PMID: 20172856
- PMCID: PMC2857135
- DOI: 10.1074/jbc.M109.067710
PB1-F2 influenza A virus protein adopts a beta-sheet conformation and forms amyloid fibers in membrane environments
Abstract
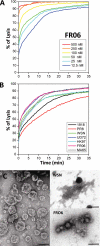
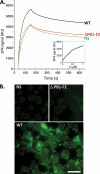
The influenza A virus PB1-F2 protein, encoded by an alternative reading frame in the PB1 polymerase gene, displays a high sequence polymorphism and is reported to contribute to viral pathogenesis in a sequence-specific manner. To gain insights into the functions of PB1-F2, the molecular structure of several PB1-F2 variants produced in Escherichia coli was investigated in different environments. Circular dichroism spectroscopy shows that all variants have a random coil secondary structure in aqueous solution. When incubated in trifluoroethanol polar solvent, all PB1-F2 variants adopt an alpha-helix-rich structure, whereas incubated in acetonitrile, a solvent of medium polarity mimicking the membrane environment, they display beta-sheet secondary structures. Incubated with asolectin liposomes and SDS micelles, PB1-F2 variants also acquire a beta-sheet structure. Dynamic light scattering revealed that the presence of beta-sheets is correlated with an oligomerization/aggregation of PB1-F2. Electron microscopy showed that PB1-F2 forms amorphous aggregates in acetonitrile. In contrast, at low concentrations of SDS, PB1-F2 variants exhibited various abilities to form fibers that were evidenced as amyloid fibers in a thioflavin T assay. Using a recombinant virus and its PB1-F2 knock-out mutant, we show that PB1-F2 also forms amyloid structures in infected cells. Functional membrane permeabilization assays revealed that the PB1-F2 variants can perforate membranes at nanomolar concentrations but with activities found to be sequence-dependent and not obviously correlated with their differential ability to form amyloid fibers. All of these observations suggest that PB1-F2 could be involved in physiological processes through different pathways, permeabilization of cellular membranes, and amyloid fiber formation.
Figures

Similar articles
-
Evolution and Virulence of Influenza A Virus Protein PB1-F2.Int J Mol Sci. 2017 Dec 29;19(1):96. doi: 10.3390/ijms19010096. Int J Mol Sci. 2017. PMID: 29286299 Free PMC article. Review.
-
Amyloid Assemblies of Influenza A Virus PB1-F2 Protein Damage Membrane and Induce Cytotoxicity.J Biol Chem. 2016 Jan 8;291(2):739-51. doi: 10.1074/jbc.M115.652917. Epub 2015 Nov 24. J Biol Chem. 2016. PMID: 26601953 Free PMC article.
-
N-terminal domain of PB1-F2 protein of influenza A virus can fold into amyloid-like oligomers and damage cholesterol and cardiolipid containing membranes.Biochem Biophys Res Commun. 2016 Aug 12;477(1):27-32. doi: 10.1016/j.bbrc.2016.06.016. Epub 2016 Jun 7. Biochem Biophys Res Commun. 2016. PMID: 27282484
-
Synchrotron Infrared and Deep UV Fluorescent Microspectroscopy Study of PB1-F2 β-Aggregated Structures in Influenza A Virus-infected Cells.J Biol Chem. 2016 Apr 22;291(17):9060-72. doi: 10.1074/jbc.M115.710533. Epub 2016 Feb 19. J Biol Chem. 2016. PMID: 26896002 Free PMC article.
-
Influenza a virus PB1-F2 protein.Acta Virol. 2007;51(2):101-8. Acta Virol. 2007. PMID: 17900216 Review.
Cited by
-
PB1-F2 attenuates virulence of highly pathogenic avian H5N1 influenza virus in chickens.PLoS One. 2014 Jun 24;9(6):e100679. doi: 10.1371/journal.pone.0100679. eCollection 2014. PLoS One. 2014. PMID: 24959667 Free PMC article.
-
Transcriptomic profiling of a chicken lung epithelial cell line (CLEC213) reveals a mitochondrial respiratory chain activity boost during influenza virus infection.PLoS One. 2017 Apr 25;12(4):e0176355. doi: 10.1371/journal.pone.0176355. eCollection 2017. PLoS One. 2017. PMID: 28441462 Free PMC article.
-
Structural Modeling of Cell Wall Peptidase CwpFM (EntFM) Reveals Distinct Intrinsically Disordered Extensions Specific to Pathogenic Bacillus cereus Strains.Toxins (Basel). 2020 Sep 14;12(9):593. doi: 10.3390/toxins12090593. Toxins (Basel). 2020. PMID: 32937845 Free PMC article.
-
Evolution and Virulence of Influenza A Virus Protein PB1-F2.Int J Mol Sci. 2017 Dec 29;19(1):96. doi: 10.3390/ijms19010096. Int J Mol Sci. 2017. PMID: 29286299 Free PMC article. Review.
-
Peptide-Induced Amyloid-Like Conformational Transitions in Proteins.Int J Pept. 2015;2015:723186. doi: 10.1155/2015/723186. Epub 2015 Sep 8. Int J Pept. 2015. PMID: 26435719 Free PMC article. Review.
References
-
- Lamb R. A., Takeda M. (2001) Nat. Med. 7, 1286–1288 - PubMed
-
- Gambotto A., Barratt-Boyes S. M., de Jong M. D., Neumann G., Kawaoka Y. (2008) Lancet 371, 1464–1475 - PubMed
-
- Garten R. J., Davis C. T., Russell C. A., Shu B., Lindstrom S., Balish A., Sessions W. M., Xu X., Skepner E., Deyde V., Okomo-Adhiambo M., Gubareva L., Barnes J., Smith C. B., Emery S. L., Hillman M. J., Rivailler P., Smagala J., de Graaf M., Burke D. F., Fouchier R. A., Pappas C., Alpuche-Aranda C. M., López-Gatell H., Olivera H., López I., Myers C. A., Faix D., Blair P. J., Yu C., Keene K. M., Dotson P. D., Jr., Boxrud D., Sambol A. R., Abid S. H., St George K., Bannerman T., Moore A. L., Stringer D. J., Blevins P., Demmler-Harrison G. J., Ginsberg M., Kriner P., Waterman S., Smole S., Guevara H. F., Belongia E. A., Clark P. A., Beatrice S. T., Donis R., Katz J., Finelli L., Bridges C. B., Shaw M., Jernigan D. B., Uyeki T. M., Smith D. J., Klimov A. I., Cox N. J. (2009) Science 325, 197–201 - PubMed
-
- Palese P. (1977) Cell 10, 1–10 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

