Nonhost resistance of barley to different fungal pathogens is associated with largely distinct, quantitative transcriptional responses
- PMID: 20172964
- PMCID: PMC2850024
- DOI: 10.1104/pp.109.151829
Nonhost resistance of barley to different fungal pathogens is associated with largely distinct, quantitative transcriptional responses
Abstract
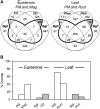
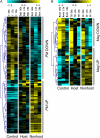
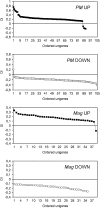
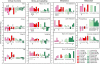
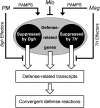
Nonhost resistance protects plants against attack by the vast majority of potential pathogens, including phytopathogenic fungi. Despite its high biological importance, the molecular architecture of nonhost resistance has remained largely unexplored. Here, we describe the transcriptional responses of one particular genotype of barley (Hordeum vulgare subsp. vulgare 'Ingrid') to three different pairs of adapted (host) and nonadapted (nonhost) isolates of fungal pathogens, which belong to the genera Blumeria (powdery mildew), Puccinia (rust), and Magnaporthe (blast). Nonhost resistance against each of these pathogens was associated with changes in transcript abundance of distinct sets of nonhost-specific genes, although general (not nonhost-associated) transcriptional responses to the different pathogens overlapped considerably. The powdery mildew- and blast-induced differences in transcript abundance between host and nonhost interactions were significantly correlated with differences between a near-isogenic pair of barley lines that carry either the Mlo wild-type allele or the mutated mlo5 allele, which mediates basal resistance to powdery mildew. Moreover, during the interactions of barley with the different host or nonhost pathogens, similar patterns of overrepresented and underrepresented functional categories of genes were found. The results suggest that nonhost resistance and basal host defense of barley are functionally related and that nonhost resistance to different fungal pathogens is associated with more robust regulation of complex but largely nonoverlapping sets of pathogen-responsive genes involved in similar metabolic or signaling pathways.
Figures

Similar articles
-
A comparative analysis of nonhost resistance across the two Triticeae crop species wheat and barley.BMC Plant Biol. 2017 Dec 4;17(1):232. doi: 10.1186/s12870-017-1178-0. BMC Plant Biol. 2017. PMID: 29202692 Free PMC article.
-
The barley (Hordeum vulgare) cellulose synthase-like D2 gene (HvCslD2) mediates penetration resistance to host-adapted and nonhost isolates of the powdery mildew fungus.New Phytol. 2016 Oct;212(2):421-33. doi: 10.1111/nph.14065. Epub 2016 Jun 28. New Phytol. 2016. PMID: 27352228
-
Barley stripe mosaic virus-induced gene silencing (BSMV-IGS) as a tool for functional analysis of barley genes potentially involved in nonhost resistance.Plant Signal Behav. 2011 Jun;6(6):867-9. doi: 10.4161/psb.6.6.15240. Epub 2011 Jun 1. Plant Signal Behav. 2011. PMID: 21586898 Free PMC article.
-
Exploring and exploiting the boundaries of host specificity using the cereal rust and mildew models.New Phytol. 2018 Apr;218(2):453-462. doi: 10.1111/nph.15044. Epub 2018 Feb 21. New Phytol. 2018. PMID: 29464724 Review.
-
Magical mystery tour: MLO proteins in plant immunity and beyond.New Phytol. 2014 Oct;204(2):273-81. doi: 10.1111/nph.12889. New Phytol. 2014. PMID: 25453131 Review.
Cited by
-
Abscisic acid negatively interferes with basal defence of barley against Magnaporthe oryzae.BMC Plant Biol. 2015 Jan 21;15:7. doi: 10.1186/s12870-014-0409-x. BMC Plant Biol. 2015. PMID: 25604965 Free PMC article.
-
A novel protein elicitor (SsCut) from Sclerotinia sclerotiorum induces multiple defense responses in plants.Plant Mol Biol. 2014 Nov;86(4-5):495-511. doi: 10.1007/s11103-014-0244-3. Epub 2014 Aug 23. Plant Mol Biol. 2014. PMID: 25149470
-
A first step toward the development of a barley NAM population and its utilization to detect QTLs conferring leaf rust seedling resistance.Theor Appl Genet. 2014 Jul;127(7):1513-25. doi: 10.1007/s00122-014-2315-x. Epub 2014 May 6. Theor Appl Genet. 2014. PMID: 24797143
-
Formae speciales of cereal powdery mildew: close or distant relatives?Mol Plant Pathol. 2014 Apr;15(3):304-14. doi: 10.1111/mpp.12093. Epub 2014 Jan 6. Mol Plant Pathol. 2014. PMID: 24286122 Free PMC article. Review.
-
Discovery of genes affecting resistance of barley to adapted and non-adapted powdery mildew fungi.Genome Biol. 2014;15(12):518. doi: 10.1186/s13059-014-0518-8. Genome Biol. 2014. PMID: 25476012 Free PMC article.
References
-
- Atienza SG, Jafary H, Niks RE. (2004) Accumulation of genes for susceptibility to Rust fungi for which barley is nearly a nonhost results in two barley lines with extreme multiple susceptibility. Planta 220: 71–79 - PubMed
-
- Breiteneder H, Mills C. (2005) Nonspecific lipid-transfer proteins in plant foods and pollens: an important allergen class. Curr Opin Allergy Clin Immunol 5: 275–279 - PubMed
-
- Broekaert WF, Cammue BPA, DeBolle MFC, Thevissen K, DeSamblanx GW, Osborn RW. (1997) Antimicrobial peptides from plants. Crit Rev Plant Sci 16: 297–323
-
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D. (2003) The molecular analysis of leaf senescence: a genomics approach. Plant Biotechnol J 1: 3–22 - PubMed
-
- Buschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergarde P, Groenendijk J, et al. (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88: 695–705 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

