Metabolic and developmental effects resulting from deletion of the citA gene encoding citrate synthase in Aspergillus nidulans
- PMID: 20173036
- PMCID: PMC2863417
- DOI: 10.1128/EC.00373-09
Metabolic and developmental effects resulting from deletion of the citA gene encoding citrate synthase in Aspergillus nidulans
Abstract
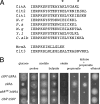
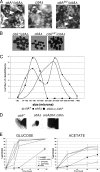
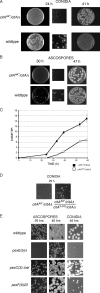
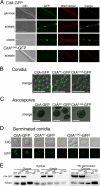
Citrate synthase is a central activity in carbon metabolism. It is required for the tricarboxylic acid (TCA) cycle, respiration, and the glyoxylate cycle. In Saccharomyces cerevisiae and Arabidopsis thaliana, there are mitochondrial and peroxisomal isoforms encoded by separate genes, while in Aspergillus nidulans, a single gene, citA, encodes a protein with predicted mitochondrial and peroxisomal targeting sequences (PTS). Deletion of citA results in poor growth on glucose but not on derepressing carbon sources, including those requiring the glyoxylate cycle. Growth on glucose is restored by a mutation in the creA carbon catabolite repressor gene. Methylcitrate synthase, required for propionyl-coenzyme A (CoA) metabolism, has previously been shown to have citrate synthase activity. We have been unable to construct the mcsADelta citADelta double mutant, and the expression of mcsA is subject to CreA-mediated carbon repression. Therefore, McsA can substitute for the loss of CitA activity. Deletion of citA does not affect conidiation or sexual development but results in delayed conidial germination as well as a complete loss of ascospores in fruiting bodies, which can be attributed to loss of meiosis. These defects are suppressed by the creA204 mutation, indicating that McsA activity can substitute for the loss of CitA. A mutation of the putative PTS1-encoding sequence in citA had no effect on carbon source utilization or development but did result in slower colony extension arising from single conidia or ascospores. CitA-green fluorescent protein (GFP) studies showed mitochondrial localization in conidia, ascospores, and hyphae. Peroxisomal localization was not detected. However, a very low and variable detection of punctate GFP fluorescence was sometimes observed in conidia germinated for 5 h when the mitochondrial targeting sequence was deleted.
Figures

Similar articles
-
Differential expression of citA gene encoding the mitochondrial citrate synthase of Aspergillus nidulans in response to developmental status and carbon sources.J Microbiol. 2010 Apr;48(2):188-98. doi: 10.1007/s12275-010-0096-8. Epub 2010 May 1. J Microbiol. 2010. PMID: 20437151
-
Cloning and characterization of the citA gene encoding the mitochondrial citrate synthase of Aspergillus nidulans.Mol Cells. 1997 Apr 30;7(2):290-5. Mol Cells. 1997. PMID: 9163747
-
Methylcitrate synthase from Aspergillus nidulans: implications for propionate as an antifungal agent.Mol Microbiol. 2000 Mar;35(5):961-73. doi: 10.1046/j.1365-2958.2000.01737.x. Mol Microbiol. 2000. PMID: 10712680
-
Blockage of methylcitrate cycle inhibits polyketide production in Aspergillus nidulans.Mol Microbiol. 2004 Apr;52(2):541-50. doi: 10.1111/j.1365-2958.2004.03994.x. Mol Microbiol. 2004. PMID: 15066039
-
Methylcitrate synthase from Aspergillus fumigatus. Propionyl-CoA affects polyketide synthesis, growth and morphology of conidia.FEBS J. 2005 Jul;272(14):3615-30. doi: 10.1111/j.1742-4658.2005.04784.x. FEBS J. 2005. PMID: 16008561
Cited by
-
Role of carnitine acetyltransferases in acetyl coenzyme A metabolism in Aspergillus nidulans.Eukaryot Cell. 2011 Apr;10(4):547-55. doi: 10.1128/EC.00295-10. Epub 2011 Feb 4. Eukaryot Cell. 2011. PMID: 21296915 Free PMC article.
-
Viral growth factor- and STAT3 signaling-dependent elevation of the TCA cycle intermediate levels during vaccinia virus infection.PLoS Pathog. 2021 Feb 2;17(2):e1009303. doi: 10.1371/journal.ppat.1009303. eCollection 2021 Feb. PLoS Pathog. 2021. PMID: 33529218 Free PMC article.
-
Differential expression of citA gene encoding the mitochondrial citrate synthase of Aspergillus nidulans in response to developmental status and carbon sources.J Microbiol. 2010 Apr;48(2):188-98. doi: 10.1007/s12275-010-0096-8. Epub 2010 May 1. J Microbiol. 2010. PMID: 20437151
-
Dynamic Regulation of Peroxisomes and Mitochondria during Fungal Development.J Fungi (Basel). 2020 Nov 20;6(4):302. doi: 10.3390/jof6040302. J Fungi (Basel). 2020. PMID: 33233491 Free PMC article. Review.
-
Reproductive competence: a recurrent logic module in eukaryotic development.Proc Biol Sci. 2013 Jul 17;280(1766):20130819. doi: 10.1098/rspb.2013.0819. Print 2013 Sep 7. Proc Biol Sci. 2013. PMID: 23864594 Free PMC article. Review.
References
-
- Armitt S., McCullough W., Roberts C. F. 1976. Analysis of acetate non-utilizing (acu) mutants in Aspergillus nidulans. J. Gen. Microbiol. 92:263–282 - PubMed
-
- Bailey C., Arst H. N., Jr 1975. Carbon catabolite repression in Aspergillus nidulans. Eur. J. Biochem. 51:573–577 - PubMed
-
- Brocard C., Hartig A. 2006. Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta 1763:1565–1573 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

