Use of ichip for high-throughput in situ cultivation of "uncultivable" microbial species
- PMID: 20173072
- PMCID: PMC2849220
- DOI: 10.1128/AEM.01754-09
Use of ichip for high-throughput in situ cultivation of "uncultivable" microbial species
Abstract
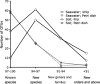
One of the oldest unresolved microbiological phenomena is why only a small fraction of the diverse microbiological population grows on artificial media. The "uncultivable" microbial majority arguably represents our planet's largest unexplored pool of biological and chemical novelty. Previously we showed that species from this pool could be grown inside diffusion chambers incubated in situ, likely because diffusion provides microorganisms with their naturally occurring growth factors. Here we utilize this approach and develop a novel high-throughput platform for parallel cultivation and isolation of previously uncultivated microbial species from a variety of environments. We have designed and tested an isolation chip (ichip) composed of several hundred miniature diffusion chambers, each inoculated with a single environmental cell. We show that microbial recovery in the ichip exceeds manyfold that afforded by standard cultivation, and the grown species are of significant phylogenetic novelty. The new method allows access to a large and diverse array of previously inaccessible microorganisms and is well suited for both fundamental and applied research.
Figures
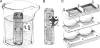
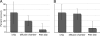

References
-
- Baltz, R. H. 2005. Antibiotics from Actinomycetes: will a renaissance follow the decline and fall? SIM News 55:186-196.
-
- Brenan, C. J. H., T. Morrison, K. Stone, T. Heitner, A. Katz, T. Kanigan, R. Hess, S.-J. Kwon, H.-C. Jung, and J.-G. Pan. 2002. A massively parallel microfluidics platform for storage and ultra high throughput screening. Proc. Soc. Photo. Opt. Instrum. Eng. 4626:560-569.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

