Sampling the N-terminal proteome of human blood
- PMID: 20173099
- PMCID: PMC2842036
- DOI: 10.1073/pnas.0914495107
Sampling the N-terminal proteome of human blood
Abstract
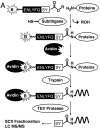
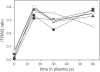
The proteomes of blood plasma and serum represent a potential gold mine of biological and diagnostic information, but challenges such as dynamic range of protein concentration have hampered efforts to unlock this resource. Here we present a method to label and isolate N-terminal peptides from human plasma and serum. This process dramatically reduces the complexity of the sample by eliminating internal peptides. We identify 772 unique N-terminal peptides in 222 proteins, ranging over six orders of magnitude in abundance. This approach is highly suited for studying natural proteolysis in plasma and serum. We find internal cleavages in plasma proteins created by endo- and exopeptidases, providing information about the activities of proteolytic enzymes in blood, which may be correlated with disease states. We also find signatures of signal peptide cleavage, coagulation and complement activation, and other known proteolytic processes, in addition to a large number of cleavages that have not been reported previously, including over 200 cleavages of blood proteins by aminopeptidases. Finally, we can identify substrates from specific proteases by exogenous addition of the protease combined with N-terminal isolation and quantitative mass spectrometry. In this way we identified proteins cleaved in human plasma by membrane-type serine protease 1, an enzyme linked to cancer progression. These studies demonstrate the utility of direct N-terminal labeling by subtiligase to identify and characterize endogenous and exogenous proteolysis in human plasma and serum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Anderson NL, Anderson NG. The human plasma proteome: History, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. - PubMed
-
- Zolotarjova N, et al. Differences among techniques for high-abundant protein depletion. Proteomics. 2005;5:3304–3313. - PubMed
-
- Cohen MP, Clements RS. Measuring glycated proteins: Clinical and methodological aspects. Diabetes Technol The. 1999;1:57–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

