Transcriptional control of the TNF gene
- PMID: 20173386
- PMCID: PMC4785889
- DOI: 10.1159/000289196
Transcriptional control of the TNF gene
Abstract
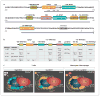
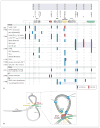
The cytokine TNF is a critical mediator of immune and inflammatory responses. The TNF gene is an immediate early gene, rapidly transcribed in a variety of cell types following exposure to a broad range of pathogens and signals of inflammation and stress. Regulation of TNF gene expression at the transcriptional level is cell type- and stimulus-specific, involving the recruitment of distinct sets of transcription factors to a compact and modular promoter region. In this review, we describe our current understanding of the mechanisms through which TNF transcription is specifically activated by a variety of extracellular stimuli in multiple cell types, including T cells, B cells, macrophages, mast cells, dendritic cells, and fibroblasts. We discuss the role of nuclear factor of activated T cells and other transcription factors and coactivators in enhanceosome formation, as well as the contradictory evidence for a role for nuclear factor kappaB as a classical activator of the TNF gene. We describe the impact of evolutionarily conserved cis-regulatory DNA motifs in the TNF locus upon TNF gene transcription, in contrast to the neutral effect of single nucleotide polymorphisms. We also assess the regulatory role of chromatin organization, epigenetic modifications, and long-range chromosomal interactions at the TNF locus.
Copyright (c) 2010 S. Karger AG, Basel.
Figures

References
-
- Aggarwal BB, Samanta A, Feldmann M. TNFα. In: Oppenheim J, Feldmann M, editors. Cytokine Reference. Orlando: Academic Press; 2000. pp. 413–434.
-
- Grivennikov SI, Kuprash DV, Liu ZG, Nedospasov SA. Intracellular signals and events activated by cytokines of the tumor necrosis factor superfamily: from simple paradigms to complex mechanisms. Int Rev Cytol. 2006;252:129–161. - PubMed
-
- Beutler B, Cerami A. Cachectin and tumor necrosis factor as two sides of the same biological coin. Nature. 1986;320:584. - PubMed
-
- Steffen M, Ottmann OG, Moore MA. Simultaneous production of tumor necrosis factor-α and lymphotoxin by normal T cells after induction with IL-2 and anti-T3. J Immunol. 1988;140:2621–2624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

