Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule
- PMID: 20174553
- PMCID: PMC2824755
- DOI: 10.1371/journal.ppat.1000776
Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule
Abstract
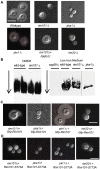
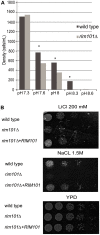
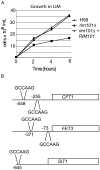
Cryptococcus neoformans is a prevalent human fungal pathogen that must survive within various tissues in order to establish a human infection. We have identified the C. neoformans Rim101 transcription factor, a highly conserved pH-response regulator in many fungal species. The rim101 multiply sign in circle mutant strain displays growth defects similar to other fungal species in the presence of alkaline pH, increased salt concentrations, and iron limitation. However, the rim101 multiply sign in circle strain is also characterized by a striking defect in capsule, an important virulence-associated phenotype. This capsular defect is likely due to alterations in polysaccharide attachment to the cell surface, not in polysaccharide biosynthesis. In contrast to many other C. neoformans capsule-defective strains, the rim101 multiply sign in circle mutant is hypervirulent in animal models of cryptococcosis. Whereas Rim101 activation in other fungal species occurs through the conserved Rim pathway, we demonstrate that C. neoformans Rim101 is also activated by the cAMP/PKA pathway. We report here that C. neoformans uses PKA and the Rim pathway to regulate the localization, activation, and processing of the Rim101 transcription factor. We also demonstrate specific host-relevant activating conditions for Rim101 cleavage, showing that C. neoformans has co-opted conserved signaling pathways to respond to the specific niche within the infected host. These results establish a novel mechanism for Rim101 activation and the integration of two conserved signaling cascades in response to host environmental conditions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

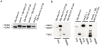
Similar articles
-
The Cryptococcus neoformans Rim101 transcription factor directly regulates genes required for adaptation to the host.Mol Cell Biol. 2014 Feb;34(4):673-84. doi: 10.1128/MCB.01359-13. Epub 2013 Dec 9. Mol Cell Biol. 2014. PMID: 24324006 Free PMC article.
-
Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses.mBio. 2013 Jan 15;4(1):e00522-12. doi: 10.1128/mBio.00522-12. mBio. 2013. PMID: 23322637 Free PMC article.
-
The Cryptococcus neoformans alkaline response pathway: identification of a novel rim pathway activator.PLoS Genet. 2015 Apr 10;11(4):e1005159. doi: 10.1371/journal.pgen.1005159. eCollection 2015 Apr. PLoS Genet. 2015. PMID: 25859664 Free PMC article.
-
The Cryptococcus neoformans capsule: a sword and a shield.Clin Microbiol Rev. 2012 Jul;25(3):387-408. doi: 10.1128/CMR.00001-12. Clin Microbiol Rev. 2012. PMID: 22763631 Free PMC article. Review.
-
The cAMP/Protein Kinase a Pathway Regulates Virulence and Adaptation to Host Conditions in Cryptococcus neoformans.Front Cell Infect Microbiol. 2019 Jun 18;9:212. doi: 10.3389/fcimb.2019.00212. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31275865 Free PMC article. Review.
Cited by
-
An encapsulation of iron homeostasis and virulence in Cryptococcus neoformans.Trends Microbiol. 2013 Sep;21(9):457-65. doi: 10.1016/j.tim.2013.05.007. Epub 2013 Jun 25. Trends Microbiol. 2013. PMID: 23810126 Free PMC article. Review.
-
Cryptococcus neoformans Cda1 and Its Chitin Deacetylase Activity Are Required for Fungal Pathogenesis.mBio. 2018 Nov 20;9(6):e02087-18. doi: 10.1128/mBio.02087-18. mBio. 2018. PMID: 30459196 Free PMC article.
-
Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans.PLoS One. 2010 Sep 2;5(9):e12503. doi: 10.1371/journal.pone.0012503. PLoS One. 2010. PMID: 20824073 Free PMC article.
-
Engineered Fluorescent Strains of Cryptococcus neoformans: a Versatile Toolbox for Studies of Host-Pathogen Interactions and Fungal Biology, Including the Viable but Nonculturable State.Microbiol Spectr. 2022 Oct 26;10(5):e0150422. doi: 10.1128/spectrum.01504-22. Epub 2022 Aug 25. Microbiol Spectr. 2022. PMID: 36005449 Free PMC article.
-
Nitrogen source-dependent capsule induction in human-pathogenic cryptococcus species.Eukaryot Cell. 2013 Nov;12(11):1439-50. doi: 10.1128/EC.00169-13. Epub 2013 Aug 23. Eukaryot Cell. 2013. PMID: 23975889 Free PMC article.
References
-
- Vartivarian SE, Anaissie EJ, Cowart RE, Sprigg HA, Tingler MJ, et al. Regulation of cryptococcal capsular polysaccharide by iron. J Infect Dis. 1993;167:186–190. - PubMed
-
- Bensen E, Martin SJ, Li M, Berman J, DA D. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol Microbiology. 2004;54:1335–1351. - PubMed
-
- Davis DA. How human pathogenic fungi sense and adapt to pH: the link to virulence. Current Opinion in Microbiology. 2009;12:365–370. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

