The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule
- PMID: 20174556
- PMCID: PMC2824758
- DOI: 10.1371/journal.ppat.1000762
The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule
Abstract
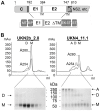
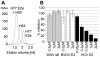
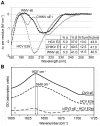
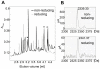
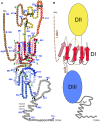
Hepatitis C virus (HCV), a major cause of chronic liver disease in humans, is the focus of intense research efforts worldwide. Yet structural data on the viral envelope glycoproteins E1 and E2 are scarce, in spite of their essential role in the viral life cycle. To obtain more information, we developed an efficient production system of recombinant E2 ectodomain (E2e), truncated immediately upstream its trans-membrane (TM) region, using Drosophila melanogaster cells. This system yields a majority of monomeric protein, which can be readily separated chromatographically from contaminating disulfide-linked aggregates. The isolated monomeric E2e reacts with a number of conformation-sensitive monoclonal antibodies, binds the soluble CD81 large external loop and efficiently inhibits infection of Huh7.5 cells by infectious HCV particles (HCVcc) in a dose-dependent manner, suggesting that it adopts a native conformation. These properties of E2e led us to experimentally determine the connectivity of its 9 disulfide bonds, which are strictly conserved across HCV genotypes. Furthermore, circular dichroism combined with infrared spectroscopy analyses revealed the secondary structure contents of E2e, indicating in particular about 28% beta-sheet, in agreement with the consensus secondary structure predictions. The disulfide connectivity pattern, together with data on the CD81 binding site and reported E2 deletion mutants, enabled the threading of the E2e polypeptide chain onto the structural template of class II fusion proteins of related flavi- and alphaviruses. The resulting model of the tertiary organization of E2 gives key information on the antigenicity determinants of the virus, maps the receptor binding site to the interface of domains I and III, and provides insight into the nature of a putative fusogenic conformational change.
Conflict of interest statement
Thomas Krey, Carlos M. Kikuti, Laurence Damier-Piolle and Félix A. Rey are listed in a patent application in the field of “HCV-derived polypeptides” that has been filed and is pending.
Figures

Similar articles
-
Construction and characterization of chimeric hepatitis C virus E2 glycoproteins: analysis of regions critical for glycoprotein aggregation and CD81 binding.J Gen Virol. 2000 Dec;81(Pt 12):2873-2883. doi: 10.1099/0022-1317-81-12-2873. J Gen Virol. 2000. PMID: 11086118
-
Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding.Virology. 2002 Jun 20;298(1):124-32. doi: 10.1006/viro.2002.1463. Virology. 2002. PMID: 12093180
-
An Optimized Hepatitis C Virus E2 Glycoprotein Core Adopts a Functional Homodimer That Efficiently Blocks Virus Entry.J Virol. 2017 Feb 14;91(5):e01668-16. doi: 10.1128/JVI.01668-16. Print 2017 Mar 1. J Virol. 2017. PMID: 28031364 Free PMC article.
-
HCV Glycoprotein Structure and Implications for B-Cell Vaccine Development.Int J Mol Sci. 2020 Sep 16;21(18):6781. doi: 10.3390/ijms21186781. Int J Mol Sci. 2020. PMID: 32947858 Free PMC article. Review.
-
Structure and Function of the Hepatitis C Virus Envelope Glycoproteins E1 and E2: Antiviral and Vaccine Targets.ACS Infect Dis. 2016 Nov 11;2(11):749-762. doi: 10.1021/acsinfecdis.6b00110. Epub 2016 Aug 16. ACS Infect Dis. 2016. PMID: 27933781 Review.
Cited by
-
Identification of Potential HCV Inhibitors Based on the Interaction of Epigallocatechin-3-Gallate with Viral Envelope Proteins.Molecules. 2021 Feb 26;26(5):1257. doi: 10.3390/molecules26051257. Molecules. 2021. PMID: 33652639 Free PMC article.
-
Proteomic approaches to analyzing hepatitis C virus biology.Proteomics. 2015 Jun;15(12):2051-65. doi: 10.1002/pmic.201500009. Epub 2015 May 5. Proteomics. 2015. PMID: 25809442 Free PMC article. Review.
-
A novel membrane fusion protein family in Flaviviridae?Trends Microbiol. 2014 Apr;22(4):176-82. doi: 10.1016/j.tim.2014.01.008. Epub 2014 Feb 23. Trends Microbiol. 2014. PMID: 24569295 Free PMC article.
-
Lectin microarray and mass spectrometric analysis of hepatitis C proteins reveals N-linked glycosylation.Medicine (Baltimore). 2018 Apr;97(15):e0208. doi: 10.1097/MD.0000000000010208. Medicine (Baltimore). 2018. PMID: 29642144 Free PMC article.
-
Hepatitis C virus E2 envelope glycoprotein core structure.Science. 2013 Nov 29;342(6162):1090-4. doi: 10.1126/science.1243876. Science. 2013. PMID: 24288331 Free PMC article.
References
-
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. - PubMed
-
- De Francesco R, Migliaccio G. Challenges and successes in developing new therapies for hepatitis C. Nature. 2005;436:953–960. - PubMed
-
- Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: The viruses and their replication. Fields Virology, Fifth edition. 2007:1101–1152.
-
- Lavie M, Goffard A, Dubuisson J. Assembly of a functional HCV glycoprotein heterodimer. Curr Issues Mol Biol. 2007;9:71–86. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

