The Nanos3-3'UTR is required for germ cell specific NANOS3 expression in mouse embryos
- PMID: 20174582
- PMCID: PMC2823788
- DOI: 10.1371/journal.pone.0009300
The Nanos3-3'UTR is required for germ cell specific NANOS3 expression in mouse embryos
Abstract
Background: The regulation of gene expression via a 3' untranslated region (UTR) plays essential roles in the discrimination of the germ cell lineage from somatic cells during embryogenesis. This is fundamental to the continuation of a species. Mouse NANOS3 is an essential protein required for the germ cell maintenance and is specifically expressed in these cells. However, the regulatory mechanisms that restrict the expression of this gene in the germ cells is largely unknown at present.
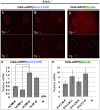
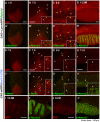
Methodology/principal findings: In our current study, we show that differences in the stability of Nanos3 mRNA between germ cells and somatic cells is brought about in a 3'UTR-dependent manner in mouse embryos. Although Nanos3 is transcribed in both cell lineages, it is efficiently translated only in the germ lineage. We also find that the translational suppression of NANOS3 in somatic cells is caused by a 3'UTR-mediated mRNA destabilizing mechanism. Surprisingly, even when under the control of the CAG promoter which induces strong ubiquitous transcription in both germ cells and somatic cells, the addition of the Nanos3-3'UTR sequence to the coding region of exogenous gene was effective in restricting protein expression in germ cells.
Conclusions/significance: Our current study thus suggests that Nanos3-3'UTR has an essential role in translational control in the mouse embryo.
Conflict of interest statement
Figures
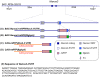
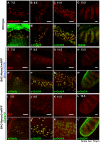
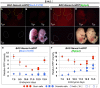
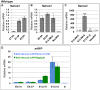
Similar articles
-
Visualization of Turbot (Scophthalmus maximus) Primordial Germ Cells in vivo Using Fluorescent Protein Mediated by the 3' Untranslated Region of nanos3 or vasa Gene.Mar Biotechnol (NY). 2019 Oct;21(5):671-682. doi: 10.1007/s10126-019-09911-z. Epub 2019 Sep 9. Mar Biotechnol (NY). 2019. PMID: 31502176
-
Conserved mechanisms for germ cell-specific localization of nanos3 transcripts in teleost species with aquaculture significance.Mar Biotechnol (NY). 2014 Jun;16(3):256-64. doi: 10.1007/s10126-013-9543-y. Epub 2013 Oct 4. Mar Biotechnol (NY). 2014. PMID: 24091820
-
Conserved elements in the nanos3 3'UTR of olive flounder are responsible for the selective retention of RNA in germ cells.Comp Biochem Physiol B Biochem Mol Biol. 2016 Aug;198:66-72. doi: 10.1016/j.cbpb.2016.04.002. Epub 2016 Apr 13. Comp Biochem Physiol B Biochem Mol Biol. 2016. PMID: 27085583
-
Primordial germ cell identification and traceability during the initial development of the Siluriformes fish Pseudopimelodus mangurus.Fish Physiol Biochem. 2022 Oct;48(5):1137-1153. doi: 10.1007/s10695-022-01106-z. Epub 2022 Aug 4. Fish Physiol Biochem. 2022. PMID: 35925505
-
Aberrant regulation of messenger RNA 3'-untranslated region in human cancer.Cell Oncol. 2007;29(1):1-17. doi: 10.1155/2007/586139. Cell Oncol. 2007. PMID: 17429137 Free PMC article. Review.
Cited by
-
Visualization of Turbot (Scophthalmus maximus) Primordial Germ Cells in vivo Using Fluorescent Protein Mediated by the 3' Untranslated Region of nanos3 or vasa Gene.Mar Biotechnol (NY). 2019 Oct;21(5):671-682. doi: 10.1007/s10126-019-09911-z. Epub 2019 Sep 9. Mar Biotechnol (NY). 2019. PMID: 31502176
-
Conserved mechanisms for germ cell-specific localization of nanos3 transcripts in teleost species with aquaculture significance.Mar Biotechnol (NY). 2014 Jun;16(3):256-64. doi: 10.1007/s10126-013-9543-y. Epub 2013 Oct 4. Mar Biotechnol (NY). 2014. PMID: 24091820
-
Both 20S and 19S proteasome components are essential for meiosis in male mice.Zool Res. 2025 Jan 18;46(1):27-40. doi: 10.24272/j.issn.2095-8137.2024.281. Zool Res. 2025. PMID: 39757018 Free PMC article.
-
Animal models for COVID-19: advances, gaps and perspectives.Signal Transduct Target Ther. 2022 Jul 7;7(1):220. doi: 10.1038/s41392-022-01087-8. Signal Transduct Target Ther. 2022. PMID: 35798699 Free PMC article. Review.
-
Molecular regulation of the mitosis/meiosis decision in multicellular organisms.Cold Spring Harb Perspect Biol. 2011 Aug 1;3(8):a002683. doi: 10.1101/cshperspect.a002683. Cold Spring Harb Perspect Biol. 2011. PMID: 21646377 Free PMC article. Review.
References
-
- D'Agostino I, Merritt C, Chen PL, Seydoux G, Subramaniam K. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev Biol. 2006;292:244–252. - PubMed
-
- Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat Rev Genet. 2003;4:626–637. - PubMed
-
- Mosquera L, Forristall C, Zhou Y, King ML. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development. 1993;117:377–386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

