HIV drug resistance surveillance using pooled pyrosequencing
- PMID: 20174661
- PMCID: PMC2822863
- DOI: 10.1371/journal.pone.0009263
HIV drug resistance surveillance using pooled pyrosequencing
Abstract
Background: Surveillance for HIV transmitted drug resistance (TDR) is performed using HIV genotype results from individual specimens. Pyrosequencing, through its massive parallel sequencing ability, can analyze large numbers of specimens simultaneously. Instead of using pyrosequencing conventionally, to sequence a population of viruses within an individual, we interrogated a single combined pool of surveillance specimens to demonstrate that it is possible to determine TDR rates in HIV protease from a population of individuals.
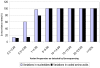
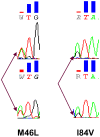
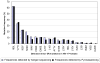
Methodology/principal findings: The protease region from 96 treatment naïve, HIV+ serum specimens was genotyped using standard Sanger sequencing method. The 462 bp protease amplicons from these specimens were pooled in equimolar concentrations and re-sequenced using the GS FLX Titanium system. The nucleotide (NT) and amino acid (AA) differences from the reference sequence, along with TDR mutations, detected by each method were compared. In the protease sequence, there were 212 nucleotide and 81 AA differences found using conventional sequencing and 345 nucleotide and 168 AA differences using pyrosequencing. All nucleotide and amino acid polymorphisms found at frequencies >/=5% in pyrosequencing were detected using both methods with the rates of variation highly correlated. Using Sanger sequencing, two TDR mutations, M46L and I84V, were each detected as mixtures at a frequency of 1.04% (1/96). These same TDR mutations were detected by pyrosequencing with a prevalence of 0.29% and 0.34% respectively. Phylogenetic analysis established that the detected low frequency mutations arose from the same single specimens that were found to contain TDR mutations by Sanger sequencing. Multiple clinical protease DR mutations present at higher frequencies were concordantly identified using both methods.
Conclusions/significance: We show that pyrosequencing pooled surveillance specimens can cost-competitively detect protease TDR mutations when compared with conventional methods. With few modifications, the method described here can be used to determine population rates of TDR in both protease and reverse transcriptase. Furthermore, this pooled pyrosequencing technique may be generalizable to other infectious agents where a survey of DR rates is required.
Conflict of interest statement
Figures

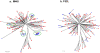

References
-
- Bennett DE. The requirement for surveillance of HIV drug resistance within antiretroviral rollout in the developing world. Curr Opin Infect Dis. 2006;19:607–614. - PubMed
-
- Bennett DE, Bertagnolio S, Sutherland D, Gilks CF. The World Health Organization's global strategy for prevention and assessment of HIV drug resistance. Antivir Ther. 2008;13(Suppl 2):1–13. - PubMed
-
- Myatt M, Bennett DE. A novel sequential sampling technique for the surveillance of transmitted HIV drug resistance by cross-sectional survey for use in low resource settings. Antivir Ther. 2008;13(Suppl 2):37–48. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

