Single molecule sensing by nanopores and nanopore devices
- PMID: 20174694
- PMCID: PMC3009472
- DOI: 10.1039/b907735a
Single molecule sensing by nanopores and nanopore devices
Abstract
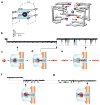
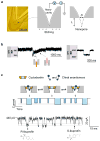
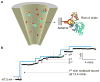
Molecular-scale pore structures, called nanopores, can be assembled by protein ion channels through genetic engineering or be artificially fabricated on solid substrates using fashion nanotechnology. When target molecules interact with the functionalized lumen of a nanopore, they characteristically block the ion pathway. The resulting conductance changes allow for identification of single molecules and quantification of target species in the mixture. In this review, we first overview nanopore-based sensory techniques that have been created for the detection of myriad biomedical targets, from metal ions, drug compounds, and cellular second messengers to proteins and DNA. Then we introduce our recent discoveries in nanopore single molecule detection: (1) using the protein nanopore to study folding/unfolding of the G-quadruplex aptamer; (2) creating a portable and durable biochip that is integrated with a single-protein pore sensor (this chip is compared with recently developed protein pore sensors based on stabilized bilayers on glass nanopore membranes and droplet interface bilayer); and (3) creating a glass nanopore-terminated probe for single-molecule DNA detection, chiral enantiomer discrimination, and identification of the bioterrorist agent ricin with an aptamer-encoded nanopore.
Figures





References
-
-
B. Hille, 2001, 3rd,
-
-
- Bayley H, Cremer PS. Nature. 2001;413:226–230. - PubMed
-
- Song LZ, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Science. 1996;274:1859–1866. - PubMed
-
- Bayley H, Cremer PS. Nature. 2001;413:226–230. - PubMed
-
- Bayley H, Jayasinghe L. Molecular Membrane Biology. 2004;21:209–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

