A rearrangement of the guanosine-binding site establishes an extended network of functional interactions in the Tetrahymena group I ribozyme active site
- PMID: 20175542
- PMCID: PMC2860537
- DOI: 10.1021/bi902200n
A rearrangement of the guanosine-binding site establishes an extended network of functional interactions in the Tetrahymena group I ribozyme active site
Abstract
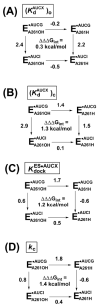
Protein enzymes appear to use extensive packing and hydrogen bonding interactions to precisely position catalytic groups within active sites. Because of their inherent backbone flexibility and limited side chain repertoire, RNA enzymes face additional challenges relative to proteins in precisely positioning substrates and catalytic groups. Here, we use the group I ribozyme to probe the existence, establishment, and functional consequences of an extended network of interactions in an RNA active site. The group I ribozyme catalyzes a site-specific attack of guanosine on an oligonucleotide substrate. We previously determined that the hydrogen bond between the exocyclic amino group of guanosine and the 2'-hydroxyl group at position A261 of the Tetrahymena group I ribozyme contributes to overall catalysis. We now use functional data, aided by double mutant cycles, to probe this hydrogen bond in the individual reaction steps of the catalytic cycle. Our results indicate that this hydrogen bond is not formed upon guanosine binding to the ribozyme but instead forms at a later stage of the catalytic cycle. Formation of this hydrogen bond is correlated with other structural rearrangements in the ribozyme's active site that are promoted by docking of the oligonucleotide substrate into the ribozyme's active site, and disruption of this interaction has deleterious consequences for the chemical transformation within the ternary complex. These results, combined with earlier results, provide insight into the nature of the multiple conformational steps used by the Tetrahymena group I ribozyme to achieve its active structure and reveal an intricate, extended network of interactions that is used to establish catalytic interactions within this RNA's active site.
Figures






Similar articles
-
An active site rearrangement within the Tetrahymena group I ribozyme releases nonproductive interactions and allows formation of catalytic interactions.RNA. 2016 Jan;22(1):32-48. doi: 10.1261/rna.053710.115. Epub 2015 Nov 13. RNA. 2016. PMID: 26567314 Free PMC article.
-
Tightening of active site interactions en route to the transition state revealed by single-atom substitution in the guanosine-binding site of the Tetrahymena group I ribozyme.J Am Chem Soc. 2011 May 25;133(20):7791-800. doi: 10.1021/ja111316y. Epub 2011 May 3. J Am Chem Soc. 2011. PMID: 21539364 Free PMC article.
-
Characterization of a local folding event of the Tetrahymena group I ribozyme: effects of oligonucleotide substrate length, pH, and temperature on the two substrate binding steps.Biochemistry. 1999 Oct 26;38(43):14192-204. doi: 10.1021/bi9914309. Biochemistry. 1999. PMID: 10571993
-
Probing the role of metal ions in RNA catalysis: kinetic and thermodynamic characterization of a metal ion interaction with the 2'-moiety of the guanosine nucleophile in the Tetrahymena group I ribozyme.Biochemistry. 1999 Aug 24;38(34):10958-75. doi: 10.1021/bi990388e. Biochemistry. 1999. PMID: 10460151
-
Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site.Mol Cell. 2004 Nov 5;16(3):351-62. doi: 10.1016/j.molcel.2004.10.003. Mol Cell. 2004. PMID: 15525509
Cited by
-
Quantitative tests of a reconstitution model for RNA folding thermodynamics and kinetics.Proc Natl Acad Sci U S A. 2017 Sep 12;114(37):E7688-E7696. doi: 10.1073/pnas.1703507114. Epub 2017 Aug 24. Proc Natl Acad Sci U S A. 2017. PMID: 28839094 Free PMC article.
-
Testing the nearest neighbor model for canonical RNA base pairs: revision of GU parameters.Biochemistry. 2012 Apr 24;51(16):3508-22. doi: 10.1021/bi3002709. Epub 2012 Apr 10. Biochemistry. 2012. PMID: 22490167 Free PMC article.
-
An active site rearrangement within the Tetrahymena group I ribozyme releases nonproductive interactions and allows formation of catalytic interactions.RNA. 2016 Jan;22(1):32-48. doi: 10.1261/rna.053710.115. Epub 2015 Nov 13. RNA. 2016. PMID: 26567314 Free PMC article.
-
Comparison of a preQ1 riboswitch aptamer in metabolite-bound and free states with implications for gene regulation.J Biol Chem. 2011 Jul 15;286(28):24626-37. doi: 10.1074/jbc.M111.230375. Epub 2011 May 18. J Biol Chem. 2011. PMID: 21592962 Free PMC article.
-
Graph Theoretical Methods and Workflows for Searching and Annotation of RNA Tertiary Base Motifs and Substructures.Int J Mol Sci. 2021 Aug 9;22(16):8553. doi: 10.3390/ijms22168553. Int J Mol Sci. 2021. PMID: 34445259 Free PMC article. Review.
References
-
- Jencks WP. Economics of enzyme catalysis. Cold Spring Harb Symp Quant Biol. 1987;52:65–73. - PubMed
-
- Ditzler MA, Aleman EA, Rueda D, Walter NG. Focus on function: single molecule RNA enzymology. Biopolymers. 2007;87:302–316. - PubMed
-
- Blount KF, Uhlenbeck OC. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct. 2005;34:415–440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

