Glioma pathophysiology: insights emerging from proteomics
- PMID: 20175778
- PMCID: PMC8094634
- DOI: 10.1111/j.1750-3639.2010.00376.x
Glioma pathophysiology: insights emerging from proteomics
Abstract
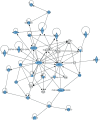
Proteomics is increasingly employed in both neurological and oncological research to provide insight into the molecular basis of disease but rarely has a coherent, novel pathophysiological insight emerged. Gliomas account for >50% of adult primary intracranial tumors, with malignant gliomas (anaplastic astrocytomas and glioblastoma multiforme) being the most common. In glioma, the application of proteomic technology has identified altered protein expression but without consistency of these alterations or their biological significance being established. A systematic review of multiple independent proteomic analyses of glioma has demonstrated alterations of 99 different proteins. Importantly 10 of the 99 proteins found differentially expressed in glioma [PHB, Hsp20, serum albumin, epidermal growth factor receptor (EGFR), EA-15, RhoGDI, APOA1, GFAP, HSP70, PDIA3] were identified in multiple publications. An assessment of protein-protein interactions between these proteins compiled using novel web-based technology, revealed a robust and cohesive network for glioblastoma. The protein network discovered (containing TP53 and RB1 at its core) compliments recent findings in genomic studies of malignant glioma. The novel perspective provided by network analysis indicates that the potential of this technology to explore crucial aspects of glioma pathophysiology can now be realized but only if the conceptual and technical limitations highlighted in this review are addressed.
Conflict of interest statement
We have no conflicts of interest.
Figures
References
-
- Albrethsen J, Knol JC, Jimenez CR (2009) Unravelling the nuclear matrix proteome. J Proteome 72:71–81. - PubMed
-
- Anderson E, Grant R, Lewis SC, Whittle IR (2008) Randomized phase III controlled trials of therapy in malignant glioma: where are we after 40 years? Br J Neurosurg 22:339–349. - PubMed
-
- Asamoto M, Cohen SM (1994) Prohibitin gene is overexpressed but not mutated in rat bladder carcinomas and cell lines. Cancer Lett 83:201–207. - PubMed
-
- Banks R, Selsby P (2003) Clinical proteomics – insights into pathologies and benefits for patients. Lancet 362:415–416. - PubMed
-
- Chakravarti A, Delaney MA, Noll E, Black PM, Loeffler JS, Muzikansky A et al (2001) Prognostic and pathologic significance of quantitative protein expression profiling in human gliomas. Clin Cancer Res 7:2387–2395. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

