Defining species specific genome differences in malaria parasites
- PMID: 20175934
- PMCID: PMC2837034
- DOI: 10.1186/1471-2164-11-128
Defining species specific genome differences in malaria parasites
Abstract
Background: In recent years a number of genome sequences for different plasmodium species have become available. This has allowed the identification of numerous conserved genes across the different species and has significantly enhanced our understanding of parasite biology. In contrast little is known about species specific differences between the different genomes partly due to the lower sequence coverage and therefore relatively poor annotation of some of the draft genomes particularly the rodent malarias parasite species.
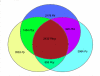
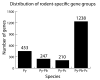
Results: To improve the current annotation and gene identification status of the draft genomes of P. berghei, P. chabaudi and P. yoelii, we performed genome-wide comparisons between these three species. Through analyses via comparative genome hybridizations using a newly designed pan-rodent array as well as in depth bioinformatics analysis, we were able to improve on the coverage of the draft rodent parasite genomes by detecting orthologous genes between these related rodent parasite species. More than 1,000 orthologs for P. yoelii were now newly associated with a P. falciparum gene. In addition to extending the current core gene set for all plasmodium species this analysis also for the first time identifies a relatively small number of genes that are unique to the primate malaria parasites while a larger gene set is uniquely conserved amongst the rodent malaria parasites.
Conclusions: These findings allow a more thorough investigation of the genes that are important for host specificity in malaria.
Figures



Similar articles
-
The phylogeny of rodent malaria parasites: simultaneous analysis across three genomes.Infect Genet Evol. 2007 Jan;7(1):74-83. doi: 10.1016/j.meegid.2006.04.005. Epub 2006 Jun 9. Infect Genet Evol. 2007. PMID: 16765106
-
Genome comparison of human and non-human malaria parasites reveals species subset-specific genes potentially linked to human disease.PLoS Comput Biol. 2011 Dec;7(12):e1002320. doi: 10.1371/journal.pcbi.1002320. Epub 2011 Dec 22. PLoS Comput Biol. 2011. PMID: 22215999 Free PMC article.
-
Genome sequences reveal divergence times of malaria parasite lineages.Parasitology. 2011 Nov;138(13):1737-49. doi: 10.1017/S0031182010001575. Epub 2010 Dec 1. Parasitology. 2011. PMID: 21118608 Free PMC article.
-
The origins, isolation, and biological characterization of rodent malaria parasites.Parasitol Int. 2022 Dec;91:102636. doi: 10.1016/j.parint.2022.102636. Epub 2022 Aug 1. Parasitol Int. 2022. PMID: 35926694 Free PMC article. Review.
-
The genome of model malaria parasites, and comparative genomics.Curr Issues Mol Biol. 2005 Jan;7(1):23-37. Curr Issues Mol Biol. 2005. PMID: 15580778 Review.
Cited by
-
Plasmodium immunomics.Int J Parasitol. 2011 Jan;41(1):3-20. doi: 10.1016/j.ijpara.2010.08.002. Epub 2010 Sep 16. Int J Parasitol. 2011. PMID: 20816843 Free PMC article. Review.
-
Integrated analysis of the Plasmodium species transcriptome.EBioMedicine. 2016 May;7:255-66. doi: 10.1016/j.ebiom.2016.04.011. Epub 2016 Apr 22. EBioMedicine. 2016. PMID: 27322479 Free PMC article.
-
Systematic CRISPR-Cas9-Mediated Modifications of Plasmodium yoelii ApiAP2 Genes Reveal Functional Insights into Parasite Development.mBio. 2017 Dec 12;8(6):e01986-17. doi: 10.1128/mBio.01986-17. mBio. 2017. PMID: 29233900 Free PMC article.
-
Plasmodium Genomics and Genetics: New Insights into Malaria Pathogenesis, Drug Resistance, Epidemiology, and Evolution.Clin Microbiol Rev. 2019 Jul 31;32(4):e00019-19. doi: 10.1128/CMR.00019-19. Print 2019 Sep 18. Clin Microbiol Rev. 2019. PMID: 31366610 Free PMC article. Review.
-
Variation in selective constraints along the Plasmodium life cycle.Infect Genet Evol. 2021 Aug;92:104908. doi: 10.1016/j.meegid.2021.104908. Epub 2021 May 8. Infect Genet Evol. 2021. PMID: 33975022 Free PMC article.
References
-
- Carter R, Diggs C. In: Parasitic Protozoa. Kreier J, editor. Vol. 3. New York: Academic Press; 1977. Plasmodia of rodents; pp. 359–465.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

