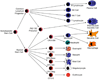
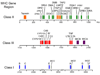
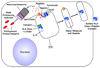
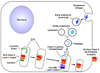
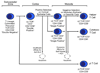
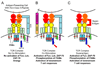
Overview of the immune response
- PMID: 20176265
- PMCID: PMC2923430
- DOI: 10.1016/j.jaci.2009.12.980
Overview of the immune response
Abstract
The immune system has evolved to protect the host from a universe of pathogenic microbes that are themselves constantly evolving. The immune system also helps the host eliminate toxic or allergenic substances that enter through mucosal surfaces. Central to the immune system's ability to mobilize a response to an invading pathogen, toxin, or allergen is its ability to distinguish self from nonself. The host uses both innate and adaptive mechanisms to detect and eliminate pathogenic microbes, and both of these mechanisms include self-nonself discrimination. This overview identifies key mechanisms used by the immune system to respond to invading microbes and other exogenous threats and identifies settings in which disturbed immune function exacerbates tissue injury.
Copyright 2010 American Academy of Allergy, Asthma & Immunology. Published by Mosby, Inc. All rights reserved.
Figures



References
-
- Hiemstra PS. The role of epithelial beta-defensins and cathelicidins in host defense of the lung. Exp Lung Res. 2007;33:537–542. - PubMed
-
- Holmskov U, Thiel S, Jensenius JC. Collectins and ficolins: humoral lectins of the innate immune defense. Annu Rev Immunol. 2003;21:547–578. - PubMed
-
- Sjoberg AP, Trouw LA, Blom AM. Complement activation and inhibition: a delicate balance. Trends Immunol. 2009;30:83–90. - PubMed
-
- Jonsson AH, Yokoyama WM. Natural killer cell tolerance licensing and other mechanisms. Adv Immunol. 2009;101:27–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

