Evolutionary dynamics of the wnt gene family: a lophotrochozoan perspective
- PMID: 20176615
- PMCID: PMC2912473
- DOI: 10.1093/molbev/msq052
Evolutionary dynamics of the wnt gene family: a lophotrochozoan perspective
Abstract
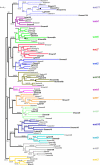
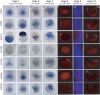
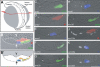
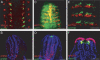
The wnt gene family encodes a set of secreted glycoproteins involved in key developmental processes, including cell fate specification and regulation of posterior growth (Cadigan KM, Nusse R. 1997. Wnt signaling: a common theme in animal development. Genes Dev. 11:3286-3305.; Martin BL, Kimelman D. 2009. Wnt signaling and the evolution of embryonic posterior development. Curr Biol. 19:R215-R219.). As for many other gene families, evidence for expansion and/or contraction of the wnt family is available from deuterostomes (e.g., echinoderms and vertebrates [Nusse R, Varmus HE. 1992. Wnt genes. Cell. 69:1073-1087.; Schubert M, Holland LZ, Holland ND, Jacobs DK. 2000. A phylogenetic tree of the Wnt genes based on all available full-length sequences, including five from the cephalochordate amphioxus. Mol Biol Evol. 17:1896-1903.; Croce JC, Wu SY, Byrum C, Xu R, Duloquin L, Wikramanayake AH, Gache C, McClay DR. 2006. A genome-wide survey of the evolutionarily conserved Wnt pathways in the sea urchin Strongylocentrotus purpuratus. Dev Biol. 300:121-131.]) and ecdysozoans (e.g., arthropods and nematodes [Eisenmann DM. 2005. Wnt signaling. WormBook. 1-17.; Bolognesi R, Farzana L, Fischer TD, Brown SJ. 2008. Multiple Wnt genes are required for segmentation in the short-germ embryo of Tribolium castaneum. Curr Biol. 18:1624-1629.]), but little is known from the third major bilaterian group, the lophotrochozoans (e.g., mollusks and annelids [Prud'homme B, Lartillot N, Balavoine G, Adoutte A, Vervoort M. 2002. Phylogenetic analysis of the Wnt gene family. Insights from lophotrochozoan members. Curr Biol. 12:1395.]). To obtain a more comprehensive scenario of the evolutionary dynamics of this gene family, we exhaustively mined wnt gene sequences from the whole genome assemblies of a mollusk (Lottia gigantea) and two annelids (Capitella teleta and Helobdella robusta) and examined them by phylogenetic, genetic linkage, intron-exon structure, and embryonic expression analyses. The 36 wnt genes obtained represent 11, 12, and 9 distinct wnt subfamilies in Lottia, Capitella, and Helobdella, respectively. Thus, two of the three analyzed lophotrochozoan genomes retained an almost complete ancestral complement of wnt genes emphasizing the importance and complexity of this gene family across metazoans. The genome of the leech Helobdella reflects significantly more dynamism than those of Lottia and Capitella, as judged by gene duplications and losses, branch length, and changes in genetic linkage. Finally, we performed a detailed expression analysis for all the Helobdella wnt genes during embryonic development. We find that, although the patterns show substantial overlap during early cleavage stages, each wnt gene has a unique expression pattern in the germinal plate and during tissue morphogenesis. Comparisons of the embryonic expression patterns of the duplicated wnt genes in Helobdella with their orthologs in Capitella reveal extensive regulatory diversification of the duplicated leech wnt genes.
Figures



References
-
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. - PubMed
-
- Blake JA, Grassle JP, Eckelbarger KJ. Capitella teleta, a new species designation for the opportunistic and experimental capitellid, Capitella sp. I, with a review of the literature for confirmed records. Zoosymposia. 2009;2:25–53.
-
- Burch JB. Regulation of GATA gene expression during vertebrate development. Semin Cell Dev Biol. 2005;16:71–81. - PubMed
-
- Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

