Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages
- PMID: 20176806
- PMCID: PMC2863586
- DOI: 10.1128/MCB.01651-09
Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages
Abstract
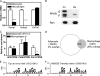
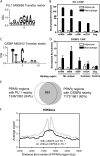
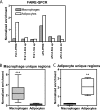
The nuclear receptor peroxisome proliferator activator receptor gamma (PPARgamma) is the target of antidiabetic thiazolidinedione drugs, which improve insulin resistance but have side effects that limit widespread use. PPARgamma is required for adipocyte differentiation, but it is also expressed in other cell types, notably macrophages, where it influences atherosclerosis, insulin resistance, and inflammation. A central question is whether PPARgamma binding in macrophages occurs at genomic locations the same as or different from those in adipocytes. Here, utilizing chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq), we demonstrate that PPARgamma cistromes in mouse adipocytes and macrophages are predominantly cell type specific. In thioglycolate-elicited macrophages, PPARgamma colocalizes with the hematopoietic transcription factor PU.1 in areas of open chromatin and histone acetylation, near a distinct set of immune genes in addition to a number of metabolic genes shared with adipocytes. In adipocytes, the macrophage-unique binding regions are marked with repressive histone modifications, typically associated with local chromatin compaction and gene silencing. PPARgamma, when introduced into preadipocytes, bound only to regions depleted of repressive histone modifications, where it increased DNA accessibility, enhanced histone acetylation, and induced gene expression. Thus, the cell specificity of PPARgamma function is regulated by cell-specific transcription factors, chromatin accessibility, and histone marks. Our data support the existence of an epigenomic hierarchy in which PPARgamma binding to cell-specific sites not marked by repressive marks opens chromatin and leads to local activation marks, including histone acetylation.
Figures




References
-
- Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837. - PubMed
-
- Bouhlel, M. A., B. Derudas, E. Rigamonti, R. Dievart, J. Brozek, S. Haulon, C. Zawadzki, B. Jude, G. Torpier, N. Marx, B. Staels, and G. Chinetti-Gbaguidi. 2007. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 6:137-143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
