Mechanism of resistance of hepatitis C virus replicons to structurally distinct cyclophilin inhibitors
- PMID: 20176894
- PMCID: PMC2863619
- DOI: 10.1128/AAC.01236-09
Mechanism of resistance of hepatitis C virus replicons to structurally distinct cyclophilin inhibitors
Abstract
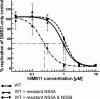
The current standard of care for hepatitis C virus (HCV) infection, pegylated alpha interferon in combination with ribavirin, has a limited response rate and adverse side effects. Drugs targeting viral proteins are in clinical development, but they suffer from the development of high viral resistance. The inhibition of cellular proteins that are essential for viral amplification is thought to have a higher barrier to the emergence of resistance. Three cyclophilin inhibitors, the cyclosporine analogs DEBIO-025, SCY635, and NIM811, have shown promising results for the treatment of HCV infection in early clinical trials. In this study, we investigated the frequency and mechanism of resistance to cyclosporine (CsA), NIM811, and a structurally unrelated cyclophilin inhibitor, SFA-1, in replicon-containing Huh7 cells. Cross-resistance between all clones was observed. NIM811-resistant clones were selected only after obtaining initial resistance to either CsA or SFA-1. The time required to select resistance against cyclophilin inhibitors was significantly longer than that required for resistance selection against viral protein inhibitors, and the achievable resistance level was substantially lower. Resistance to cyclophilin inhibitors was mediated by amino acid substitutions in NS3, NS5A, and NS5B, with NS5A mutations conferring the majority of resistance. Mutation D320E in NS5A mediated most of the resistance conferred by NS5A. Taken together, the results indicate that there is a very low frequency and level of resistance to cyclophilin-binding drugs mediated by amino acid substitutions in three viral proteins. The interaction of cyclophilin with NS5A seems to be the most critical, since the NS5A mutations have the largest impact on resistance.
Figures
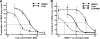
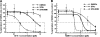

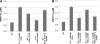
Similar articles
-
Cyclosporine inhibits a direct interaction between cyclophilins and hepatitis C NS5A.PLoS One. 2010 Mar 23;5(3):e9815. doi: 10.1371/journal.pone.0009815. PLoS One. 2010. PMID: 20352119 Free PMC article.
-
Resistance to cyclosporin A derives from mutations in hepatitis C virus nonstructural proteins.Biochem Biophys Res Commun. 2014 May 23;448(1):56-62. doi: 10.1016/j.bbrc.2014.04.053. Epub 2014 Apr 19. Biochem Biophys Res Commun. 2014. PMID: 24751518
-
Identification of cellular and viral factors related to anti-hepatitis C virus activity of cyclophilin inhibitor.Cancer Sci. 2009 Oct;100(10):1943-50. doi: 10.1111/j.1349-7006.2009.01263.x. Epub 2009 Jun 26. Cancer Sci. 2009. PMID: 19659609 Free PMC article.
-
Cyclophilin inhibitors for the treatment of HCV infection.Curr Opin Investig Drugs. 2010 Aug;11(8):911-8. Curr Opin Investig Drugs. 2010. PMID: 20721833 Review.
-
Cyclophilin inhibitors for hepatitis C therapy.Clin Liver Dis. 2013 Feb;17(1):129-39. doi: 10.1016/j.cld.2012.09.008. Clin Liver Dis. 2013. PMID: 23177289 Review.
Cited by
-
Current and emerging antiviral treatments for hepatitis C infection.Br J Clin Pharmacol. 2013 Apr;75(4):931-43. doi: 10.1111/j.1365-2125.2012.04419.x. Br J Clin Pharmacol. 2013. PMID: 22882367 Free PMC article. Review.
-
Multiple mutations in hepatitis C virus NS5A domain II are required to confer a significant level of resistance to alisporivir.Antimicrob Agents Chemother. 2012 Oct;56(10):5113-21. doi: 10.1128/AAC.00919-12. Epub 2012 Jul 16. Antimicrob Agents Chemother. 2012. PMID: 22802259 Free PMC article.
-
DEB025 (Alisporivir) inhibits hepatitis C virus replication by preventing a cyclophilin A induced cis-trans isomerisation in domain II of NS5A.PLoS One. 2010 Oct 27;5(10):e13687. doi: 10.1371/journal.pone.0013687. PLoS One. 2010. PMID: 21060866 Free PMC article.
-
Cyclophilin inhibitors as a novel HCV therapy.Viruses. 2010 Aug;2(8):1621-1634. doi: 10.3390/v2081621. Epub 2010 Aug 5. Viruses. 2010. PMID: 21994697 Free PMC article.
-
New frontiers of HCV therapy in HIV/HCV co-infection.Curr HIV/AIDS Rep. 2010 Aug;7(3):117-26. doi: 10.1007/s11904-010-0051-7. Curr HIV/AIDS Rep. 2010. PMID: 20563864
References
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- Chisari, F. V. 2005. Unscrambling hepatitis C virus-host interactions. Nature 436:930-932. - PubMed
-
- DeFrancesco, R., and G. Migliaccio. 2005. Challenges and successes in developing new therapies for hepatitis C. Nature 436:953-960. - PubMed
-
- Feld, J. J., and J. H. Hoofnagle. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436:967-972. - PubMed
-
- Fernandes, F., D. S. Poole, S. Hoover, R. Middleton, A.-C. Andrei, and J. Gerstner. 2007. Sensitivity of hepatitis C virus to cyclosporine A depends on nonstructural proteins NS5A and NS5B. Hepatology 46:1026-1033. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

