In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435
- PMID: 20176898
- PMCID: PMC2863659
- DOI: 10.1128/AAC.01452-09
In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435
Abstract
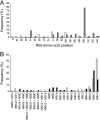
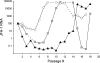
TMC435 is a small-molecule inhibitor of the NS3/4A serine protease of hepatitis C virus (HCV) currently in phase 2 development. The in vitro resistance profile of TMC435 was characterized by selection experiments with HCV genotype 1 replicon cells and the genotype 2a JFH-1 system. In 80% (86/109) of the sequences from genotype 1 replicon cells analyzed, a mutation at NS3 residue D168 was observed, with changes to V or A being the most frequent. Mutations at NS3 positions 43, 80, 155, and 156, alone or in combination, were also identified. A transient replicon assay confirmed the relevance of these positions for TMC435 inhibitory activity. The change in the 50% effective concentrations (EC(50)s) observed for replicons with mutations at position 168 ranged from <10-fold for those with the D168G or D168N mutation to approximately 2,000-fold for those with the D168V or D168I mutation, compared to the EC(50) for the wild type. Of the positions identified, mutations at residue Q80 had the least impact on the activity of TMC435 (<10-fold change in EC(50)s), while greater effects were observed for some replicons with mutations at positions 43, 155, and 156. TMC435 remained active against replicons with the specific mutations observed after in vitro or in vivo exposure to telaprevir or boceprevir, including most replicons with changes at positions 36, 54, and 170 (<3-fold change in EC(50)s). Replicons carrying mutations affecting the activity of TMC435 remained fully susceptible to alpha interferon and NS5A and NS5B inhibitors. Finally, combinations of TMC435 with alpha interferon and NS5B polymerase inhibitors prevented the formation of drug-resistant replicon colonies.
Figures

Similar articles
-
In Vitro Antiviral Activity and Resistance Profile of the Next-Generation Hepatitis C Virus NS3/4A Protease Inhibitor Glecaprevir.Antimicrob Agents Chemother. 2017 Dec 21;62(1):e01620-17. doi: 10.1128/AAC.01620-17. Print 2018 Jan. Antimicrob Agents Chemother. 2017. PMID: 29084747 Free PMC article.
-
Virologic response and characterisation of HCV genotype 2-6 in patients receiving TMC435 monotherapy (study TMC435-C202).J Hepatol. 2013 Mar;58(3):445-51. doi: 10.1016/j.jhep.2012.10.028. Epub 2012 Nov 7. J Hepatol. 2013. PMID: 23142061
-
In Vitro Activity of Simeprevir against Hepatitis C Virus Genotype 1 Clinical Isolates and Its Correlation with NS3 Sequence and Site-Directed Mutants.Antimicrob Agents Chemother. 2015 Dec;59(12):7548-57. doi: 10.1128/AAC.01444-15. Epub 2015 Sep 21. Antimicrob Agents Chemother. 2015. PMID: 26392483 Free PMC article.
-
[Significance of hepatitis C virus baseline polymorphism during the antiviral therapy].Orv Hetil. 2015 May 24;156(21):849-54. doi: 10.1556/650.2015.30180. Orv Hetil. 2015. PMID: 26038992 Review. Hungarian.
-
TMC-435, an NS3/4A protease inhibitor for the treatment of HCV infection.Curr Opin Investig Drugs. 2009 Aug;10(8):871-81. Curr Opin Investig Drugs. 2009. PMID: 19649931 Review.
Cited by
-
Dynamics of resistance mutations to NS3 protease inhibitors in a cohort of Brazilian patients chronically infected with hepatitis C virus (genotype 1) treated with pegylated interferon and ribavirin: a prospective longitudinal study.Virol J. 2013 Feb 14;10:57. doi: 10.1186/1743-422X-10-57. Virol J. 2013. PMID: 23409973 Free PMC article.
-
Hepatitis C viral entry inhibitors prolong viral suppression by replication inhibitors in persistently-infected Huh7 cultures.PLoS One. 2013 Jun 3;8(6):e65273. doi: 10.1371/journal.pone.0065273. Print 2013. PLoS One. 2013. PMID: 23755208 Free PMC article.
-
Hepatitis C virus NS3 mutations in haemophiliacs.Haemophilia. 2014 Sep;20(5):659-65. doi: 10.1111/hae.12420. Epub 2014 Apr 3. Haemophilia. 2014. PMID: 24697920 Free PMC article.
-
Hepatitis C virus molecular evolution: transmission, disease progression and antiviral therapy.World J Gastroenterol. 2014 Nov 21;20(43):15992-6013. doi: 10.3748/wjg.v20.i43.15992. World J Gastroenterol. 2014. PMID: 25473152 Free PMC article. Review.
-
Discovery of MK-1220: A Macrocyclic Inhibitor of Hepatitis C Virus NS3/4A Protease with Improved Preclinical Plasma Exposure.ACS Med Chem Lett. 2011 Jan 12;2(3):207-12. doi: 10.1021/ml1002426. eCollection 2011 Mar 10. ACS Med Chem Lett. 2011. PMID: 24900304 Free PMC article.
References
-
- Burns, C. J., A. M. Del Vecchio, T. R. Bailey, B. A. Kulkarni, T. H. Faitg, S. R. Sherk, C. W. Blackledge, D. J. Rys, T. A. Lessen, J. Swestock, Y. Deng, T. J. Nitz, J. A. Reinhardt, H. Feng, and A. K. Saha. May 2004. Benzofuran compounds, compositions and methods for treatment and prophylaxis of hepatitis C viral infections and associated diseases. PCT patent WO2004041201.
-
- Chan, L., O. Pereira, T. J. Reddy, S. K. Das, C. Poisson, M. Courchesne, M. Proulx, A. Siddiqui, C. G. Yannopoulos, N. Nguyen-Ba, C. Roy, D. Nasturica, C. Moinet, R. Bethell, M. Hamel, L. L'Heureux, M. David, O. Nicolas, P. Courtemanche-Asselin, S. Brunette, D. Bilimoria, and J. Bédard. 2004. Discovery of thiophene-2-carboxylic acids as potent inhibitors of HCV NS5B polymerase and HCV subgenomic RNA replication. Part 2. Tertiary amides. Bioorg. Med. Chem. Lett. 14:797-800. - PubMed
-
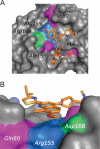
- Cummings, M. D., J. Lindberg, T.-I. Lin, H. de Kock, O. Lenz, E. Lilja, S. Felländer, V. Baraznenok, S. Nyström, M. Nilsson, L. Vrang, M. Edlund, Å. Rosenquist, B. Samuelsson, P. Raboisson, and K. Simmen. 2010. Induced fit binding of the noncovalent macrocyclic inhibitor TMC435 to its HCV NS3/NS4A protease target. Angew. Chem. Int. Ed. Engl. 49:1652-1655. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

