High-throughput identification of protein localization dependency networks
- PMID: 20176934
- PMCID: PMC2842071
- DOI: 10.1073/pnas.1000846107
High-throughput identification of protein localization dependency networks
Abstract
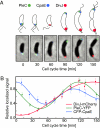
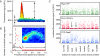
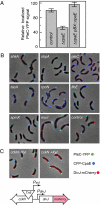
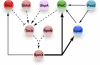
Bacterial cells are highly organized with many protein complexes and DNA loci dynamically positioned to distinct subcellular sites over the course of a cell cycle. Such dynamic protein localization is essential for polar organelle development, establishment of asymmetry, and chromosome replication during the Caulobacter crescentus cell cycle. We used a fluorescence microscopy screen optimized for high-throughput to find strains with anomalous temporal or spatial protein localization patterns in transposon-generated mutant libraries. Automated image acquisition and analysis allowed us to identify genes that affect the localization of two polar cell cycle histidine kinases, PleC and DivJ, and the pole-specific pili protein CpaE, each tagged with a different fluorescent marker in a single strain. Four metrics characterizing the observed localization patterns of each of the three labeled proteins were extracted for hundreds of cell images from each of 854 mapped mutant strains. Using cluster analysis of the resulting set of 12-element vectors for each of these strains, we identified 52 strains with mutations that affected the localization pattern of the three tagged proteins. This information, combined with quantitative localization data from epitasis experiments, also identified all previously known proteins affecting such localization. These studies provide insights into factors affecting the PleC/DivJ localization network and into regulatory links between the localization of the pili assembly protein CpaE and the kinase localization pathway. Our high-throughput screening methodology can be adapted readily to any sequenced bacterial species, opening the potential for databases of localization regulatory networks across species, and investigation of localization network phylogenies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Protein sequences and cellular factors required for polar localization of a histidine kinase in Caulobacter crescentus.J Bacteriol. 2002 Nov;184(21):6037-49. doi: 10.1128/JB.184.21.6037-6049.2002. J Bacteriol. 2002. PMID: 12374838 Free PMC article.
-
The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator.Mol Microbiol. 2003 Feb;47(4):929-41. doi: 10.1046/j.1365-2958.2003.03349.x. Mol Microbiol. 2003. PMID: 12581350
-
Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins.Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13831-6. doi: 10.1073/pnas.182411999. Epub 2002 Oct 7. Proc Natl Acad Sci U S A. 2002. PMID: 12370432 Free PMC article.
-
Dynamic localization of proteins and DNA during a bacterial cell cycle.Nat Rev Mol Cell Biol. 2002 Mar;3(3):167-76. doi: 10.1038/nrm758. Nat Rev Mol Cell Biol. 2002. PMID: 11994737 Review.
-
Protein localization during the Caulobacter crescentus cell cycle.Curr Opin Microbiol. 1998 Dec;1(6):636-42. doi: 10.1016/s1369-5274(98)80108-2. Curr Opin Microbiol. 1998. PMID: 10066543 Review.
Cited by
-
The caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor.J Bacteriol. 2010 Oct;192(19):4847-58. doi: 10.1128/JB.00607-10. Epub 2010 Aug 6. J Bacteriol. 2010. PMID: 20693330 Free PMC article.
-
The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation.Mol Cell. 2011 Oct 21;44(2):252-64. doi: 10.1016/j.molcel.2011.09.010. Mol Cell. 2011. PMID: 22017872 Free PMC article.
-
Automated quantitative live cell fluorescence microscopy.Cold Spring Harb Perspect Biol. 2010 Aug;2(8):a000455. doi: 10.1101/cshperspect.a000455. Epub 2010 Jun 30. Cold Spring Harb Perspect Biol. 2010. PMID: 20591990 Free PMC article. Review.
-
The essential genome of a bacterium.Mol Syst Biol. 2011 Aug 30;7:528. doi: 10.1038/msb.2011.58. Mol Syst Biol. 2011. PMID: 21878915 Free PMC article.
-
Dynamical Localization of DivL and PleC in the Asymmetric Division Cycle of Caulobacter crescentus: A Theoretical Investigation of Alternative Models.PLoS Comput Biol. 2015 Jul 17;11(7):e1004348. doi: 10.1371/journal.pcbi.1004348. eCollection 2015 Jul. PLoS Comput Biol. 2015. PMID: 26186202 Free PMC article.
References
-
- Shapiro L, McAdams HH, Losick R. Generating and exploiting polarity in bacteria. Science. 2002;298:1942–1946. - PubMed
-
- Gordon GS, et al. Chromosome and low copy plasmid segregation in E. coli: Visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. - PubMed
-
- Arigoni F, Pogliano K, Webb CD, Stragier P, Losick R. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science. 1995;270:637–640. - PubMed
-
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

