Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme
- PMID: 20176960
- PMCID: PMC2842065
- DOI: 10.1073/pnas.0914443107
Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme
Abstract
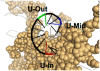
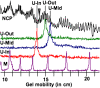
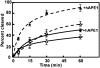
Histones play a crucial role in the organization of DNA in the nucleus, but their presence can prevent interactions with DNA binding proteins responsible for repair of DNA damage. Uracil is an abundant mutagenic lesion recognized by uracil DNA glycosylase (UDG) in the first step of base excision repair (BER). In nucleosome core particles (NCPs), we find substantial differences in UDG-directed cleavage at uracils rotationally positioned toward (U-In) or away from (U-Out) the histone core, or midway between these orientations (U-Mid). Whereas U-Out NCPs show a cleavage rate just below that of naked DNA, U-In and U-Mid NCPs have markedly slower rates of cleavage. Crosslinking of U-In DNA to histones in NCPs yields a greater reduction in cleavage rate but, surprisingly, yields a higher rate of cleavage in U-Out NCPs compared with uncrosslinked NCPs. Moreover, the next enzyme in BER, APE1, stimulates the activity of human UDG in U-Out NCPs, suggesting these enzymes interact on the surface of histones in orientations accessible to UDG. These data indicate that the activity of UDG likely requires "trapping" transiently exposed states arising from the rotational dynamics of DNA on histones.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Histone variants H3.3 and H2A.Z/H3.3 facilitate excision of uracil from nucleosome core particles.DNA Repair (Amst). 2022 Aug;116:103355. doi: 10.1016/j.dnarep.2022.103355. Epub 2022 Jun 12. DNA Repair (Amst). 2022. PMID: 35717761 Free PMC article.
-
The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes.J Biol Chem. 2013 May 10;288(19):13863-75. doi: 10.1074/jbc.M112.441444. Epub 2013 Mar 29. J Biol Chem. 2013. PMID: 23543741 Free PMC article.
-
Site-specific Acetylation of Histone H3 Decreases Polymerase β Activity on Nucleosome Core Particles in Vitro.J Biol Chem. 2016 May 20;291(21):11434-45. doi: 10.1074/jbc.M116.725788. Epub 2016 Mar 31. J Biol Chem. 2016. PMID: 27033702 Free PMC article.
-
Accessing DNA damage in chromatin: Preparing the chromatin landscape for base excision repair.DNA Repair (Amst). 2015 Aug;32:113-119. doi: 10.1016/j.dnarep.2015.04.021. Epub 2015 May 2. DNA Repair (Amst). 2015. PMID: 25957487 Free PMC article. Review.
-
Lessons learned from structural results on uracil-DNA glycosylase.Mutat Res. 2000 Aug 30;460(3-4):183-99. doi: 10.1016/s0921-8777(00)00026-4. Mutat Res. 2000. PMID: 10946228 Review.
Cited by
-
Rules of engagement for base excision repair in chromatin.J Cell Physiol. 2013 Feb;228(2):258-66. doi: 10.1002/jcp.24134. J Cell Physiol. 2013. PMID: 22718094 Free PMC article. Review.
-
Reduced Nuclease Activity of Apurinic/Apyrimidinic Endonuclease (APE1) Variants on Nucleosomes: IDENTIFICATION OF ACCESS RESIDUES.J Biol Chem. 2015 Aug 21;290(34):21067-21075. doi: 10.1074/jbc.M115.665547. Epub 2015 Jul 1. J Biol Chem. 2015. PMID: 26134573 Free PMC article.
-
Nucleosomes determine their own patch size in base excision repair.Sci Rep. 2016 Jun 6;6:27122. doi: 10.1038/srep27122. Sci Rep. 2016. PMID: 27265863 Free PMC article.
-
Nucleotide Excision Repair and Impact of Site-Specific 5',8-Cyclopurine and Bulky DNA Lesions on the Physical Properties of Nucleosomes.Biochemistry. 2019 Feb 12;58(6):561-574. doi: 10.1021/acs.biochem.8b01066. Epub 2019 Jan 7. Biochemistry. 2019. PMID: 30570250 Free PMC article.
-
Histone variants H3.3 and H2A.Z/H3.3 facilitate excision of uracil from nucleosome core particles.DNA Repair (Amst). 2022 Aug;116:103355. doi: 10.1016/j.dnarep.2022.103355. Epub 2022 Jun 12. DNA Repair (Amst). 2022. PMID: 35717761 Free PMC article.
References
-
- Friedberg EC, et al. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006.
-
- Seeberg E, Eide L, Bjørås M. The base excision repair pathway. Trends Biochem Sci. 1995;20:391–397. - PubMed
-
- Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451:39–51. - PubMed
-
- Dianov GL, Parsons JL. Co-ordination of DNA single strand break repair. DNA Repair (Amst) 2007;6:454–460. - PubMed
-
- Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

