Specificity landscapes of DNA binding molecules elucidate biological function
- PMID: 20176964
- PMCID: PMC2842033
- DOI: 10.1073/pnas.0914023107
Specificity landscapes of DNA binding molecules elucidate biological function
Abstract
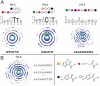
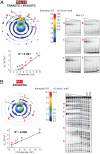
Evaluating the specificity spectra of DNA binding molecules is a nontrivial challenge that hinders the ability to decipher gene regulatory networks or engineer molecules that act on genomes. Here we compare the DNA sequence specificities for different classes of proteins and engineered DNA binding molecules across the entire sequence space. These high-content data are visualized and interpreted using an interactive "specificity landscape" which simultaneously displays the affinity and specificity of a million-plus DNA sequences. Contrary to expectation, specificity landscapes reveal that synthetic DNA ligands match, and often surpass, the specificities of eukaryotic DNA binding proteins. The landscapes also identify differential specificity constraints imposed by diverse structural folds of natural and synthetic DNA binders. Importantly, the sequence context of a binding site significantly influences binding energetics, and utilizing the full contextual information permits greater accuracy in annotating regulatory elements within a given genome. Assigning such context-dependent binding values to every DNA sequence across the genome yields predictive genome-wide binding landscapes (genomescapes). A genomescape of a synthetic DNA binding molecule provided insight into its differential regulatory activity in cultured cells. The approach we describe will accelerate the creation of precision-tailored DNA therapeutics and uncover principles that govern sequence-specificity of DNA binding molecules.
Conflict of interest statement
The authors declare a conflict of interest. A.Z.A. is a founder/proprietor and C.L.W. and M.S.O. are part-time employees of VistaMotif, and M.S.O. owns Invitrogen stock.
Figures



References
-
- Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. - PubMed
-
- Hauschild KE, Carlson CD, Donato LJ, Moretti R, Ansari AZ. Transcription Factors. In: Begley T, editor. Wiley Encyclopedia of Chemical Biology. Vol. 4. New York: John Wiley & Sons, Inc; 2008. pp. 566–584.
-
- Ptashne M, Gann A. Genes & Signals. New York: Cold Springs Harbor Laboratory Press; 2002.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

