ParA2, a Vibrio cholerae chromosome partitioning protein, forms left-handed helical filaments on DNA
- PMID: 20176965
- PMCID: PMC2842031
- DOI: 10.1073/pnas.0913060107
ParA2, a Vibrio cholerae chromosome partitioning protein, forms left-handed helical filaments on DNA
Abstract
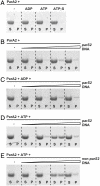
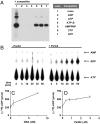
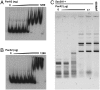
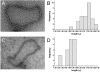
Most bacterial chromosomes contain homologs of plasmid partitioning (par) loci. These loci encode ATPases called ParA that are thought to contribute to the mechanical force required for chromosome and plasmid segregation. In Vibrio cholerae, the chromosome II (chrII) par locus is essential for chrII segregation. Here, we found that purified ParA2 had ATPase activities comparable to other ParA homologs, but, unlike many other ParA homologs, did not form high molecular weight complexes in the presence of ATP alone. Instead, formation of high molecular weight ParA2 polymers required DNA. Electron microscopy and three-dimensional reconstruction revealed that ParA2 formed bipolar helical filaments on double-stranded DNA in a sequence-independent manner. These filaments had a distinct change in pitch when ParA2 was polymerized in the presence of ATP versus in the absence of a nucleotide cofactor. Fitting a crystal structure of a ParA protein into our filament reconstruction showed how a dimer of ParA2 binds the DNA. The filaments formed with ATP are left-handed, but surprisingly these filaments exert no topological changes on the right-handed B-DNA to which they are bound. The stoichiometry of binding is one dimer for every eight base pairs, and this determines the geometry of the ParA2 filaments with 4.4 dimers per 120 A pitch left-handed turn. Our findings will be critical for understanding how ParA proteins function in plasmid and chromosome segregation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Kinetic principles of ParA2-ATP cycling guide dynamic subcellular localizations in Vibrio cholerae.Nucleic Acids Res. 2023 Jun 23;51(11):5603-5620. doi: 10.1093/nar/gkad321. Nucleic Acids Res. 2023. PMID: 37140034 Free PMC article.
-
The structure of the bacterial DNA segregation ATPase filament reveals the conformational plasticity of ParA upon DNA binding.Nat Commun. 2021 Aug 27;12(1):5166. doi: 10.1038/s41467-021-25429-2. Nat Commun. 2021. PMID: 34453062 Free PMC article.
-
Molecular Anatomy of ParA-ParA and ParA-ParB Interactions during Plasmid Partitioning.J Biol Chem. 2015 Jul 24;290(30):18782-95. doi: 10.1074/jbc.M115.649632. Epub 2015 Jun 8. J Biol Chem. 2015. PMID: 26055701 Free PMC article.
-
ParA ATPases can move and position DNA and subcellular structures.Curr Opin Microbiol. 2011 Dec;14(6):712-8. doi: 10.1016/j.mib.2011.09.008. Epub 2011 Sep 29. Curr Opin Microbiol. 2011. PMID: 21963112 Review.
-
Plasmid and chromosome partitioning: surprises from phylogeny.Mol Microbiol. 2000 Aug;37(3):455-66. doi: 10.1046/j.1365-2958.2000.01975.x. Mol Microbiol. 2000. PMID: 10931339 Review.
Cited by
-
Insight into centromere-binding properties of ParB proteins: a secondary binding motif is essential for bacterial genome maintenance.Nucleic Acids Res. 2013 Mar 1;41(5):3094-103. doi: 10.1093/nar/gkt018. Epub 2013 Jan 23. Nucleic Acids Res. 2013. PMID: 23345617 Free PMC article.
-
MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition.Proc Natl Acad Sci U S A. 2013 Jul 2;110(27):E2441-50. doi: 10.1073/pnas.1309499110. Epub 2013 Jun 17. Proc Natl Acad Sci U S A. 2013. PMID: 23776210 Free PMC article.
-
Breaking and restoring the hydrophobic core of a centromere-binding protein.J Biol Chem. 2015 Apr 3;290(14):9273-83. doi: 10.1074/jbc.M115.638148. Epub 2015 Feb 23. J Biol Chem. 2015. PMID: 25713077 Free PMC article.
-
Rules and Exceptions: The Role of Chromosomal ParB in DNA Segregation and Other Cellular Processes.Microorganisms. 2020 Jan 11;8(1):0. doi: 10.3390/microorganisms8010105. Microorganisms. 2020. PMID: 31940850 Free PMC article. Review.
-
Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB.PLoS One. 2011;6(7):e21921. doi: 10.1371/journal.pone.0021921. Epub 2011 Jul 8. PLoS One. 2011. PMID: 21760923 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

