Amplification of the gene for isoleucyl-tRNA synthetase facilitates adaptation to the fitness cost of mupirocin resistance in Salmonella enterica
- PMID: 20176977
- PMCID: PMC2870965
- DOI: 10.1534/genetics.109.113514
Amplification of the gene for isoleucyl-tRNA synthetase facilitates adaptation to the fitness cost of mupirocin resistance in Salmonella enterica
Abstract
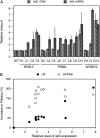
Mutations that cause resistance to antibiotics in bacteria often reduce growth rate by impairing some essential cellular function. This growth impairment is expected to counterselect resistant organisms from natural populations following discontinuation of antibiotic therapy. Unfortunately (for disease control) bacteria adapt and improve their growth rate, often without losing antibiotic resistance. This adaptation process was studied in mupirocin-resistant (Mup(R)) strains of Salmonella enterica. Mupirocin (Mup) is an isoleucyl-adenylate analog that inhibits the essential enzyme, isoleucyl-tRNA synthetase (IleRS). Mutations causing Mup(R) alter IleRS and reduce growth rate. Fitness is restored by any of 23 secondary IleRS amino acid substitutions, 60% of which leave resistance unaffected. Evidence that increased expression of the original mutant ileS gene (Mup(R)) also improves fitness while maintaining resistance is presented. Expression can be increased by amplification of the ileS gene (more copies) or mutations that improve the ileS promoter (more transcription). Some adapted strains show both ileS amplification and an improved promoter. This suggests a process of adaptation initiated by common amplifications and followed by later acquisition of rare point mutations. Finally, a point mutation in one copy relaxes selection and allows loss of defective ileS copies. This sequence of events is demonstrated experimentally. A better understanding of adaptation can explain why antibiotic resistance persists in bacterial populations and may help identify drugs that are least subject to this problem.
Figures
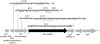

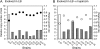
Similar articles
-
Multiple mechanisms to ameliorate the fitness burden of mupirocin resistance in Salmonella typhimurium.Mol Microbiol. 2007 May;64(4):1038-48. doi: 10.1111/j.1365-2958.2007.05713.x. Mol Microbiol. 2007. PMID: 17501926
-
Relationship of protein structure of isoleucyl-tRNA synthetase with pseudomonic acid resistance of Escherichia coli. A proposed mode of action of pseudomonic acid as an inhibitor of isoleucyl-tRNA synthetase.J Biol Chem. 1994 Sep 30;269(39):24304-9. J Biol Chem. 1994. PMID: 7929087
-
The isoleucyl-tRNA synthetase mutation V588F conferring mupirocin resistance in glycopeptide-intermediate Staphylococcus aureus is not associated with a significant fitness burden.J Antimicrob Chemother. 2004 Jan;53(1):102-4. doi: 10.1093/jac/dkh020. Epub 2003 Dec 4. J Antimicrob Chemother. 2004. PMID: 14657089
-
Exploring mechanisms of mupirocin resistance and hyper-resistance.Biochem Soc Trans. 2024 Jun 26;52(3):1109-1120. doi: 10.1042/BST20230581. Biochem Soc Trans. 2024. PMID: 38884776 Review.
-
Clinical relevance of mupirocin resistance in Staphylococcus aureus.J Hosp Infect. 2013 Dec;85(4):249-56. doi: 10.1016/j.jhin.2013.09.006. Epub 2013 Sep 21. J Hosp Infect. 2013. PMID: 24144552 Review.
Cited by
-
Antibiotic resistance mediated by gene amplifications.NPJ Antimicrob Resist. 2024 Nov 6;2(1):35. doi: 10.1038/s44259-024-00052-5. NPJ Antimicrob Resist. 2024. PMID: 39843582 Free PMC article. Review.
-
Gene duplication as a mechanism of genomic adaptation to a changing environment.Proc Biol Sci. 2012 Dec 22;279(1749):5048-57. doi: 10.1098/rspb.2012.1108. Epub 2012 Sep 12. Proc Biol Sci. 2012. PMID: 22977152 Free PMC article. Review.
-
Large-scale duplication events underpin population-level flexibility in tRNA gene copy number in Pseudomonas fluorescens SBW25.Nucleic Acids Res. 2024 Mar 21;52(5):2446-2462. doi: 10.1093/nar/gkae049. Nucleic Acids Res. 2024. PMID: 38296823 Free PMC article.
-
Multilevel gene expression changes in lineages containing adaptive copy number variants.bioRxiv [Preprint]. 2024 Jul 9:2023.10.20.563336. doi: 10.1101/2023.10.20.563336. bioRxiv. 2024. Update in: Mol Biol Evol. 2025 Feb 03;42(2):msaf005. doi: 10.1093/molbev/msaf005. PMID: 37961325 Free PMC article. Updated. Preprint.
-
Challenges of antibacterial discovery.Clin Microbiol Rev. 2011 Jan;24(1):71-109. doi: 10.1128/CMR.00030-10. Clin Microbiol Rev. 2011. PMID: 21233508 Free PMC article. Review.
References
-
- Andersson, D. I., 2003. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol. 6 452–456. - PubMed
-
- Andersson, D. I., 2006. The biological cost of mutational antibiotic resistance: Any practical conclusions? Curr. Opin. Microbiol. 9 461–465. - PubMed
-
- Andersson, D. I., and D. Hughes, 2009. Gene amplification and adaptive evolution in bacteria. Annu. Rev. Genet. 43 167–195. - PubMed
-
- Andersson, D. I., and B. R. Levin, 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2 489–493. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

