Fission yeast Hsk1 (Cdc7) kinase is required after replication initiation for induced mutagenesis and proper response to DNA alkylation damage
- PMID: 20176980
- PMCID: PMC2870973
- DOI: 10.1534/genetics.109.112284
Fission yeast Hsk1 (Cdc7) kinase is required after replication initiation for induced mutagenesis and proper response to DNA alkylation damage
Abstract
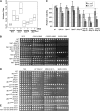
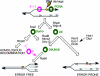
Genome stability in fission yeast requires the conserved S-phase kinase Hsk1 (Cdc7) and its partner Dfp1 (Dbf4). In addition to their established function in the initiation of DNA replication, we show that these proteins are important in maintaining genome integrity later in S phase and G2. hsk1 cells suffer increased rates of mitotic recombination and require recombination proteins for survival. Both hsk1 and dfp1 mutants are acutely sensitive to alkylation damage yet defective in induced mutagenesis. Hsk1 and Dfp1 are associated with the chromatin even after S phase, and normal response to MMS damage correlates with the maintenance of intact Dfp1 on chromatin. A screen for MMS-sensitive mutants identified a novel truncation allele, rad35 (dfp1-(1-519)), as well as alleles of other damage-associated genes. Although Hsk1-Dfp1 functions with the Swi1-Swi3 fork protection complex, it also acts independently of the FPC to promote DNA repair. We conclude that Hsk1-Dfp1 kinase functions post-initiation to maintain replication fork stability, an activity potentially mediated by the C terminus of Dfp1.
Figures




Similar articles
-
Hsk1-Dfp1/Him1, the Cdc7-Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex.J Biol Chem. 2005 Dec 30;280(52):42536-42. doi: 10.1074/jbc.M510575200. Epub 2005 Oct 31. J Biol Chem. 2005. PMID: 16263721
-
A conserved domain of Schizosaccharomyces pombe dfp1(+) is uniquely required for chromosome stability following alkylation damage during S phase.Mol Cell Biol. 2002 Jul;22(13):4477-90. doi: 10.1128/MCB.22.13.4477-4490.2002. Mol Cell Biol. 2002. PMID: 12052858 Free PMC article.
-
Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage.Mol Cell Biol. 2005 Apr;25(7):2770-84. doi: 10.1128/MCB.25.7.2770-2784.2005. Mol Cell Biol. 2005. PMID: 15767681 Free PMC article.
-
Regulation of chromosome dynamics by Hsk1/Cdc7 kinase.Biochem Soc Trans. 2013 Dec;41(6):1712-9. doi: 10.1042/BST20130217. Biochem Soc Trans. 2013. PMID: 24256280 Review.
-
Cdc7 kinases (DDKs) and checkpoint responses: lessons from two yeasts.Mutat Res. 2003 Nov 27;532(1-2):21-7. doi: 10.1016/j.mrfmmm.2003.08.007. Mutat Res. 2003. PMID: 14643426 Review.
Cited by
-
Phosphorylated Rad18 directs DNA polymerase η to sites of stalled replication.J Cell Biol. 2010 Nov 29;191(5):953-66. doi: 10.1083/jcb.201006043. Epub 2010 Nov 22. J Cell Biol. 2010. PMID: 21098111 Free PMC article.
-
The C-terminus of S. pombe DDK subunit Dfp1 is required for meiosis-specific transcription and cohesin cleavage.Biol Open. 2013 Jun 11;2(7):728-38. doi: 10.1242/bio.20135173. Print 2013 Jul 15. Biol Open. 2013. PMID: 23862021 Free PMC article.
-
ATR-Chk1-APC/CCdh1-dependent stabilization of Cdc7-ASK (Dbf4) kinase is required for DNA lesion bypass under replication stress.Genes Dev. 2013 Nov 15;27(22):2459-72. doi: 10.1101/gad.224568.113. Genes Dev. 2013. PMID: 24240236 Free PMC article.
-
Diploidy confers genomic instability in Schizosaccharomyces pombe.bioRxiv [Preprint]. 2025 Mar 10:2025.02.04.636513. doi: 10.1101/2025.02.04.636513. bioRxiv. 2025. Update in: Genetics. 2025 Jun 4;230(2):iyaf078. doi: 10.1093/genetics/iyaf078. PMID: 39975392 Free PMC article. Updated. Preprint.
-
Eukaryotic DNA polymerase ζ.DNA Repair (Amst). 2015 May;29:47-55. doi: 10.1016/j.dnarep.2015.02.012. Epub 2015 Feb 19. DNA Repair (Amst). 2015. PMID: 25737057 Free PMC article. Review.
References
-
- Andersen, P. L., F. Xu and W. Xiao, 2008. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 18 162–173. - PubMed
-
- Bailis, J. M., P. Bernard, R. Antonelli, R. Allshire and S. L. Forsburg, 2003. Hsk1/Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat. Cell Biol. 5 1111–1116. - PubMed
-
- Barbour, L., and W. Xiao, 2003. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat. Res. 532 137–155. - PubMed
-
- Bernard, P., J. F. Maure, J. F. Partridge, S. Genier, J. P. Javerzat et al., 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294 2539–2542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

