Human carboxymethylenebutenolidase as a bioactivating hydrolase of olmesartan medoxomil in liver and intestine
- PMID: 20177059
- PMCID: PMC2852926
- DOI: 10.1074/jbc.M109.072629
Human carboxymethylenebutenolidase as a bioactivating hydrolase of olmesartan medoxomil in liver and intestine
Abstract
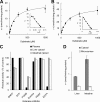
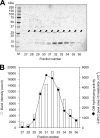
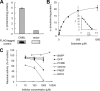
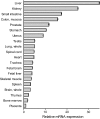
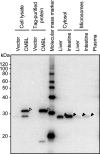
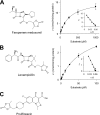
Olmesartan medoxomil (OM) is a prodrug type angiotensin II type 1 receptor antagonist widely prescribed as an antihypertensive agent. Herein, we describe the identification and characterization of the OM bioactivating enzyme that hydrolyzes the prodrug and converts to its pharmacologically active metabolite olmesartan in human liver and intestine. The protein was purified from human liver cytosol by successive column chromatography and was identified by mass spectrometry to be a carboxymethylenebutenolidase (CMBL) homolog. Human CMBL, whose endogenous function has still not been reported, is a human homolog of Pseudomonas dienelactone hydrolase involved in the bacterial halocatechol degradation pathway. The ubiquitous expression of human CMBL gene transcript in various tissues was observed. The recombinant human CMBL expressed in mammalian cells was clearly shown to activate OM. By comparing the enzyme kinetics and chemical inhibition properties between the recombinant protein and human tissue preparations, CMBL was demonstrated to be the primary OM bioactivating enzyme in the liver and intestine. The recombinant CMBL also converted other prodrugs having the same ester structure as OM, faropenem medoxomil and lenampicillin, to their active metabolites. CMBL exhibited a unique sensitivity to chemical inhibitors, thus, being distinguishable from other known esterases. Site-directed mutagenesis on the putative active residue Cys(132) of the recombinant CMBL caused a drastic reduction of the OM-hydrolyzing activity. We report for the first time that CMBL serves as a key enzyme in the bioactivation of OM, hydrolyzing the ester bond of the prodrug type xenobiotics.
Figures


Similar articles
-
Interindividual variability of carboxymethylenebutenolidase homolog, a novel olmesartan medoxomil hydrolase, in the human liver and intestine.Drug Metab Dispos. 2013 May;41(5):1156-62. doi: 10.1124/dmd.113.051482. Epub 2013 Mar 7. Drug Metab Dispos. 2013. PMID: 23471504
-
Different hydrolases involved in bioactivation of prodrug-type angiotensin receptor blockers: carboxymethylenebutenolidase and carboxylesterase 1.Drug Metab Dispos. 2013 Nov;41(11):1888-95. doi: 10.1124/dmd.113.053595. Epub 2013 Aug 14. Drug Metab Dispos. 2013. PMID: 23946449
-
Paraoxonase 1 as a major bioactivating hydrolase for olmesartan medoxomil in human blood circulation: molecular identification and contribution to plasma metabolism.Drug Metab Dispos. 2012 Feb;40(2):374-80. doi: 10.1124/dmd.111.041475. Epub 2011 Nov 15. Drug Metab Dispos. 2012. PMID: 22086979
-
The new oral angiotensin II antagonist olmesartan medoxomil: a concise overview.J Hum Hypertens. 2002 May;16 Suppl 2:S13-6. doi: 10.1038/sj.jhh.1001391. J Hum Hypertens. 2002. PMID: 11967728 Review.
-
Clinical efficacy of olmesartan medoxomil.J Hypertens Suppl. 2003 May;21(2):S43-6. doi: 10.1097/00004872-200305002-00008. J Hypertens Suppl. 2003. PMID: 12929907 Review.
Cited by
-
Characterization of a novel oxidase from Thelonectria discophora SANK 18292 involved in nectrisine biosynthesis.AMB Express. 2016 Mar;6(1):6. doi: 10.1186/s13568-016-0176-1. Epub 2016 Jan 20. AMB Express. 2016. PMID: 26786316 Free PMC article.
-
Differential protein expression profiles of cyst fluid from papillary thyroid carcinoma and benign thyroid lesions.PLoS One. 2015 May 15;10(5):e0126472. doi: 10.1371/journal.pone.0126472. eCollection 2015. PLoS One. 2015. PMID: 25978681 Free PMC article.
-
Xenobiotic Metabolism and Gut Microbiomes.PLoS One. 2016 Oct 3;11(10):e0163099. doi: 10.1371/journal.pone.0163099. eCollection 2016. PLoS One. 2016. PMID: 27695034 Free PMC article.
-
Impact of chronic intermittent hypoxia on the long non-coding RNA and mRNA expression profiles in myocardial infarction.J Cell Mol Med. 2021 Jan;25(1):421-433. doi: 10.1111/jcmm.16097. Epub 2020 Nov 20. J Cell Mol Med. 2021. PMID: 33215878 Free PMC article.
-
Genetic association with overall survival of taxane-treated lung cancer patients - a genome-wide association study in human lymphoblastoid cell lines followed by a clinical association study.BMC Cancer. 2012 Sep 24;12:422. doi: 10.1186/1471-2407-12-422. BMC Cancer. 2012. PMID: 23006423 Free PMC article.
References
-
- Testa B., Mayer J. M. (2003) Hydrolysis in Drug and Prodrug Metabolism. Chemistry, Biochemistry, and Enzymology, 1st Ed., pp. 1–3, Wiley-VCH, Zürich, Switzerland
-
- Ettmayer P., Amidon G. L., Clement B., Testa B. (2004) J. Med. Chem. 47, 2393–2404 - PubMed
-
- Beaumont K., Webster R., Gardner I., Dack K. (2003) Curr. Drug Metab. 4, 461–485 - PubMed
-
- Testa B. (2004) Biochem. Pharmacol. 68, 2097–2106 - PubMed
-
- Satoh T., Hosokawa M. (2006) Chem. Biol. Interact. 162, 195–211 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

