Ataxia telangiectasia mutated (ATM) inhibition transforms human mammary gland epithelial cells
- PMID: 20177072
- PMCID: PMC2857118
- DOI: 10.1074/jbc.M109.078360
Ataxia telangiectasia mutated (ATM) inhibition transforms human mammary gland epithelial cells
Abstract
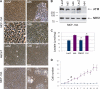
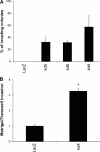
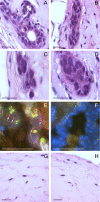
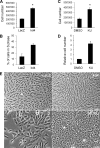
Carriers of mutations in the cell cycle checkpoint protein kinase ataxia telangiectasia mutated (ATM), which represent 1-2% of the general population, have an increased risk of breast cancer. However, experimental evidence that ATM deficiency contributes to human breast carcinogenesis is lacking. We report here that in MCF-10A and MCF-12A cells, which are well established normal human mammary gland epithelial cell models, partial or almost complete stable ATM silencing or pharmacological inhibition resulted in cellular transformation, genomic instability, and formation of dysplastic lesions in NOD/SCID mice. These effects did not require the activity of exogenous DNA-damaging agents and were preceded by an unsuspected and striking increase in cell proliferation also observed in primary human mammary gland epithelial cells. Increased proliferation correlated with a dramatic, transient, and proteasome-dependent reduction of p21(WAF1/CIP1) and p27(KIP1) protein levels, whereas little or no effect was observed on p21(WAF1/CIP1) or p27(KIP1) mRNAs. p21(WAF1/CIP1) silencing also increased MCF-10A cell proliferation, thus identifying p21(WAF1/CIP1) down-regulation as a mediator of the proliferative effect of ATM inhibition. Our findings provide the first experimental evidence that ATM is a human breast tumor suppressor. In addition, they mirror the sensitivity of ATM tumor suppressor function and unveil a new mechanism by which ATM might prevent human breast tumorigenesis, namely a direct inhibitory effect on the basal proliferation of normal mammary epithelial cells.
Figures


Similar articles
-
DNA protein kinase-dependent G2 checkpoint revealed following knockdown of ataxia-telangiectasia mutated in human mammary epithelial cells.Cancer Res. 2008 Jan 1;68(1):89-97. doi: 10.1158/0008-5472.CAN-07-0675. Cancer Res. 2008. PMID: 18172300 Free PMC article.
-
Recruitment of ataxia-telangiectasia mutated to the p21(waf1) promoter by ZBP-89 plays a role in mucosal protection.Gastroenterology. 2006 Sep;131(3):841-52. doi: 10.1053/j.gastro.2006.06.014. Gastroenterology. 2006. PMID: 16952553
-
The p53-p21WAF1 checkpoint pathway plays a protective role in preventing DNA rereplication induced by abrogation of FOXF1 function.Cell Signal. 2012 Jan;24(1):316-24. doi: 10.1016/j.cellsig.2011.09.017. Epub 2011 Sep 22. Cell Signal. 2012. PMID: 21964066 Free PMC article.
-
Antioxidants suppress lymphoma and increase longevity in Atm-deficient mice.J Nutr. 2007 Jan;137(1 Suppl):229S-232S. doi: 10.1093/jn/137.1.229S. J Nutr. 2007. PMID: 17182831 Review.
-
AMP-activated protein kinase (AMPK) beyond metabolism: a novel genomic stress sensor participating in the DNA damage response pathway.Cancer Biol Ther. 2014 Feb;15(2):156-69. doi: 10.4161/cbt.26726. Epub 2013 Nov 1. Cancer Biol Ther. 2014. PMID: 24100703 Free PMC article. Review.
Cited by
-
Ataxia-telangiectasia mutated (ATM) silencing promotes neuroblastoma progression through a MYCN independent mechanism.Oncotarget. 2015 Jul 30;6(21):18558-76. doi: 10.18632/oncotarget.4061. Oncotarget. 2015. PMID: 26053094 Free PMC article.
-
SIM2s directed Parkin-mediated mitophagy promotes mammary epithelial cell differentiation.Cell Death Differ. 2023 Jun;30(6):1472-1487. doi: 10.1038/s41418-023-01146-9. Epub 2023 Mar 25. Cell Death Differ. 2023. PMID: 36966227 Free PMC article.
-
ATR- and ATM-Mediated DNA Damage Response Is Dependent on Excision Repair Assembly during G1 but Not in S Phase of Cell Cycle.PLoS One. 2016 Jul 21;11(7):e0159344. doi: 10.1371/journal.pone.0159344. eCollection 2016. PLoS One. 2016. PMID: 27442013 Free PMC article.
-
Genotoxic exposure during juvenile growth of mammary gland depletes stem cell activity and inhibits Wnt signaling.PLoS One. 2012;7(11):e49902. doi: 10.1371/journal.pone.0049902. Epub 2012 Nov 21. PLoS One. 2012. PMID: 23185480 Free PMC article.
-
Aluminium chloride promotes tumorigenesis and metastasis in normal murine mammary gland epithelial cells.Int J Cancer. 2016 Dec 15;139(12):2781-2790. doi: 10.1002/ijc.30393. Epub 2016 Sep 7. Int J Cancer. 2016. PMID: 27541736 Free PMC article.
References
-
- Lavin M. F. (2008) Nat. Rev. Mol. Cell Biol. 9, 759–769 - PubMed
-
- Renwick A., Thompson D., Seal S., Kelly P., Chagtai T., Ahmed M., North B., Jayatilake H., Barfoot R., Spanova K., McGuffog L., Evans D. G., Eccles D., Easton D. F., Stratton M. R., Rahman N. (2006) Nat. Genet. 38, 873–875 - PubMed
-
- Hall J. (2005) Cancer Lett. 227, 105–114 - PubMed
-
- Weil M. M., Kittrell F. S., Yu Y., McCarthy M., Zabriskie R. C., Ullrich R. L. (2001) Oncogene 20, 4409–4411 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

