Got target? Computational methods for microRNA target prediction and their extension
- PMID: 20177143
- PMCID: PMC2859323
- DOI: 10.3858/emm.2010.42.4.032
Got target? Computational methods for microRNA target prediction and their extension
Abstract
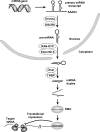
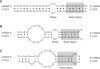
MicroRNAs (miRNAs) are a class of small RNAs of 19-23 nucleotides that regulate gene expression through target mRNA degradation or translational gene silencing. The miRNAs are reported to be involved in many biological processes, and the discovery of miRNAs has been provided great impacts on computational biology as well as traditional biology. Most miRNA-associated computational methods comprise the prediction of miRNA genes and their targets, and increasing numbers of computational algorithms and web-based resources are being developed to fulfill the need of scientists performing miRNA research. Here we summarize the rules to predict miRNA targets and introduce some computational algorithms that have been developed for miRNA target prediction and the application of the methods. In addition, the issue of target gene validation in an experimental way will be discussed.
Figures
Similar articles
-
Computational prediction of microRNA targets.Methods Mol Biol. 2010;667:283-95. doi: 10.1007/978-1-60761-811-9_19. Methods Mol Biol. 2010. PMID: 20827541
-
Computational analysis of microRNA targets in Caenorhabditis elegans.Gene. 2006 Jan 3;365:2-10. doi: 10.1016/j.gene.2005.09.035. Epub 2005 Dec 13. Gene. 2006. PMID: 16356665
-
Prediction of human microRNA targets.Methods Mol Biol. 2006;342:101-13. doi: 10.1385/1-59745-123-1:101. Methods Mol Biol. 2006. PMID: 16957370 Review.
-
Identification of new stress-induced microRNA and their targets in wheat using computational approach.Plant Signal Behav. 2013 May;8(5):e23932. doi: 10.4161/psb.23932. Epub 2013 Mar 19. Plant Signal Behav. 2013. PMID: 23511197 Free PMC article.
-
Computational identification of microRNAs and their targets.Birth Defects Res C Embryo Today. 2006 Jun;78(2):118-28. doi: 10.1002/bdrc.20067. Birth Defects Res C Embryo Today. 2006. PMID: 16847881 Review.
Cited by
-
Systems Drug Design for Muscle Invasive Bladder Cancer and Advanced Bladder Cancer by Genome-Wide Microarray Data and Deep Learning Method with Drug Design Specifications.Int J Mol Sci. 2022 Nov 10;23(22):13869. doi: 10.3390/ijms232213869. Int J Mol Sci. 2022. PMID: 36430344 Free PMC article.
-
OrbId: Origin-based identification of microRNA targets.Mob Genet Elements. 2012 Jul 1;2(4):184-192. doi: 10.4161/mge.21617. Mob Genet Elements. 2012. PMID: 23087843 Free PMC article.
-
Computational prediction of intronic microRNA targets using host gene expression reveals novel regulatory mechanisms.PLoS One. 2011;6(6):e19312. doi: 10.1371/journal.pone.0019312. Epub 2011 Jun 9. PLoS One. 2011. PMID: 21694770 Free PMC article.
-
Delivery and targeting of miRNAs for treating liver fibrosis.Pharm Res. 2015 Feb;32(2):341-61. doi: 10.1007/s11095-014-1497-x. Epub 2014 Sep 4. Pharm Res. 2015. PMID: 25186440 Review.
-
Correlation between microRNA-421 expression level and prognosis of gastric cancer.Int J Clin Exp Pathol. 2015 Nov 1;8(11):15128-32. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26823855 Free PMC article.
References
-
- Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–3055. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579:5904–5910. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
