Predicting protein crystallization propensity from protein sequence
- PMID: 20177794
- PMCID: PMC3366497
- DOI: 10.1007/s10969-010-9080-0
Predicting protein crystallization propensity from protein sequence
Abstract
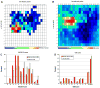
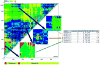
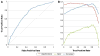
The high-throughput structure determination pipelines developed by structural genomics programs offer a unique opportunity for data mining. One important question is how protein properties derived from a primary sequence correlate with the protein's propensity to yield X-ray quality crystals (crystallizability) and 3D X-ray structures. A set of protein properties were computed for over 1,300 proteins that expressed well but were insoluble, and for approximately 720 unique proteins that resulted in X-ray structures. The correlation of the protein's iso-electric point and grand average hydropathy (GRAVY) with crystallizability was analyzed for full length and domain constructs of protein targets. In a second step, several additional properties that can be calculated from the protein sequence were added and evaluated. Using statistical analyses we have identified a set of the attributes correlating with a protein's propensity to crystallize and implemented a Support Vector Machine (SVM) classifier based on these. We have created applications to analyze and provide optimal boundary information for query sequences and to visualize the data. These tools are available via the web site http://bioinformatics.anl.gov/cgi-bin/tools/pdpredictor .
Figures


References
-
- Gao X, et al. High-throughput limited proteolysis/mass spectrometry for protein domain elucidation. J Struct Funct Genomics. 2005;6(2–3):129–134. - PubMed
-
- Koth CM, et al. Use of limited proteolysis to identify protein domains suitable for structural analysis. Methods Enzymol. 2003;368:77–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

