Evolution of spliceosomal introns following endosymbiotic gene transfer
- PMID: 20178587
- PMCID: PMC2834692
- DOI: 10.1186/1471-2148-10-57
Evolution of spliceosomal introns following endosymbiotic gene transfer
Abstract
Background: Spliceosomal introns are an ancient, widespread hallmark of eukaryotic genomes. Despite much research, many questions regarding the origin and evolution of spliceosomal introns remain unsolved, partly due to the difficulty of inferring ancestral gene structures. We circumvent this problem by using genes originated by endosymbiotic gene transfer, in which an intron-less structure at the time of the transfer can be assumed.
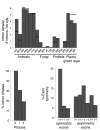
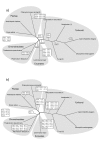
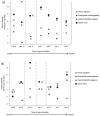
Results: By comparing the exon-intron structures of 64 mitochondrial-derived genes that were transferred to the nucleus at different evolutionary periods, we can trace the history of intron gains in different eukaryotic lineages. Our results show that the intron density of genes transferred relatively recently to the nuclear genome is similar to that of genes originated by more ancient transfers, indicating that gene structure can be rapidly shaped by intron gain after the integration of the gene into the genome and that this process is mainly determined by forces acting specifically on each lineage. We analyze 12 cases of mitochondrial-derived genes that have been transferred to the nucleus independently in more than one lineage.
Conclusions: Remarkably, the proportion of shared intron positions that were gained independently in homologous genes is similar to that proportion observed in genes that were transferred prior to the speciation event and whose shared intron positions might be due to vertical inheritance. A particular case of parallel intron gain in the nad7 gene is discussed in more detail.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

