Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus
- PMID: 20178741
- PMCID: PMC2828953
- DOI: 10.1016/j.cell.2010.01.029
Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus
Abstract
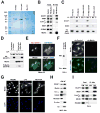
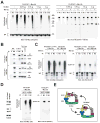
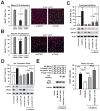
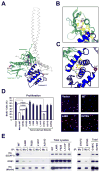
Current models imply that the FERM domain protein Merlin, encoded by the tumor suppressor NF2, inhibits mitogenic signaling at or near the plasma membrane. Here, we show that the closed, growth-inhibitory form of Merlin accumulates in the nucleus, binds to the E3 ubiquitin ligase CRL4(DCAF1), and suppresses its activity. Depletion of DCAF1 blocks the promitogenic effect of inactivation of Merlin. Conversely, enforced expression of a Merlin-insensitive mutant of DCAF1 counteracts the antimitogenic effect of Merlin. Re-expression of Merlin and silencing of DCAF1 implement a similar, tumor-suppressive program of gene expression. Tumor-derived mutations invariably disrupt Merlin's ability to interact with or inhibit CRL4(DCAF1). Finally, depletion of DCAF1 inhibits the hyperproliferation of Schwannoma cells from NF2 patients and suppresses the oncogenic potential of Merlin-deficient tumor cell lines. We propose that Merlin suppresses tumorigenesis by translocating to the nucleus to inhibit CRL4(DCAF1).
2010 Elsevier Inc. All rights reserved.
Figures


References
-
- Ahronowitz I, Xin W, Kiely R, Sims K, MacCollin M, Nunes FP. Mutational spectrum of the NF2 gene: a meta-analysis of 12 years of research and diagnostic laboratory findings. Hum Mutat. 2007;28:1–12. - PubMed
-
- Ammoun S, Flaiz C, Ristic N, Schuldt J, Hanemann CO. Dissecting and targeting the growth factor-dependent and growth factor-independent extracellular signal-regulated kinase pathway in human schwannoma. Cancer Res. 2008;68:5236–5245. - PubMed
-
- Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. - PubMed
-
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. - PubMed
-
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

